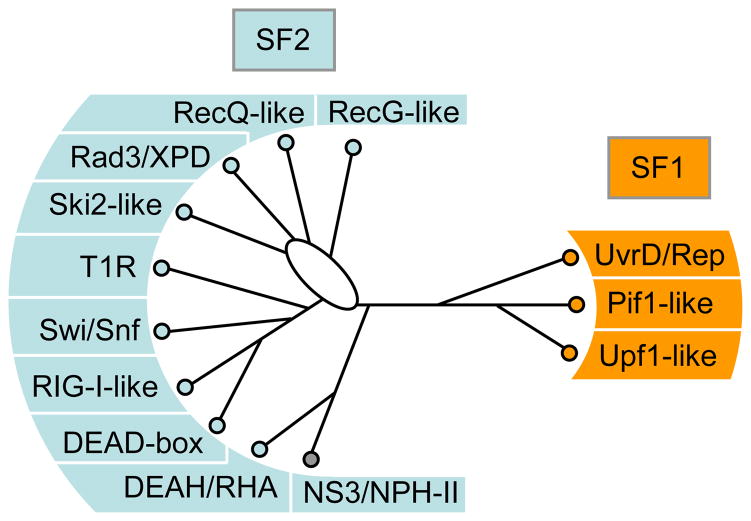
Figure 1. The Families of the SF1 and SF2 helicases.
Schematic, unrooted cladogram showing the three identified families of the SF1 (right), and the 9 families and one group of the SF2 (left). A comprehensive list of proteins in each family in human, S.cerevisiae, and E.coli is given in the Supplementary Tables ST1 and ST2. Families were identified by a combination of phylogenetic analysis of the alignment of all SF1 and SF2 proteins from human, S.cerevisiae, E.coli, and selected viruses (Suppl. Fig. S1), and scoring for presence or absence of distinct sequence features and characteristic domain organization (for more detail, see Suppl. Figs. S1–3) and corresponding captions). Assignment of proteins to a family was highly robust. The phylogenetic relationships between the families were more ambiguous and tree topologies varied for some branches according to method and parameters used for generating the trees (Suppl. Fig. S2). The cladogram shown represents an average of multiple trees generated by different means (Suppl. Fig. S2). Branch lengths are not to scale. The oval indicates significant uncertainty in the tree topology in this region. Families were named according to names in use, or according to prominent members. Families named after a single protein were termed – like (i.e., Ski2-like). Non-standard abbreviations are as follows: T1R – type 1 restriction enzymes, RHA – RNA helicase A.