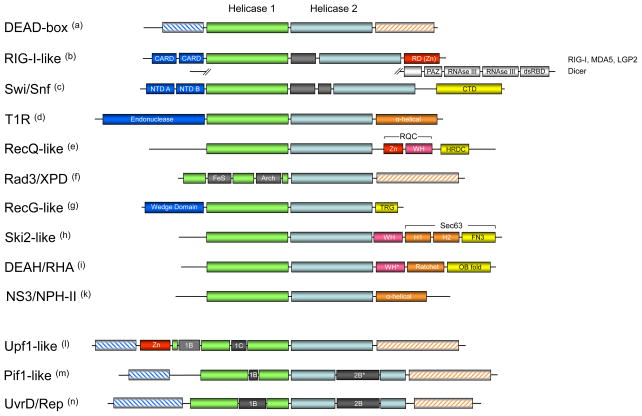
Figure 3. Domain organization of SF2 and SF1 helicase families and groups.
Domains are not to scale. (a) C- and N-termini of DEAD-box proteins include RRMs, Zn-fingers, tudor domains and others [26]. (b) The family-typical domain inserted between the helicase domains is shown in grey. RIG-I-like proteins vary in their terminal domains [70]. Prominent RIG-I-like proteins are shown, Mph1p/FancM-related proteins are not shown. (CARD – caspase recruitment domain, RD – regulatory domain, a Zn-binding domain [67], PAZ – PIWI, Argonaute, Zwille, dsRBD – double strand RNA binding domain) (c) Domain organization of bacterial RapA [93]. Inserted domain between the helicase core domains is family-typical (Suppl. Fig. 1) (NTD – N-terminal domain, CTD – C terminal domain). (d) Domain organization of EcoR124 [80]. (e) RecQ-like proteins feature multiple C and N-terminal domains including exonuclease domains. Shown are the most conserved features of the RecQ-like family [94]. (Zn – Zn finger domain, WH – winged helix domain, HRDC – Helicase and RNAseD-like C terminal domain, RQC – RecQ C-terminal domain) (f) Rad3/XPD proteins also feature diverse C and N-termini. The domains inserted in helicase domain 1 are family-typical (FeS – iron-sulfur cluster, Arch – Arch domain, a structural domain, see Fig. 4) (g) Domain organization in RecG-like protein is largely conserved, but PriA has a non-typical Zn finger inserted in the helicase domain 2 [66] (TRG – translocation by RecG) (h) Domain organization of Hel 308 [52]. The organization of the C-terminal domains is conserved in Brr2 [95,96]. (WH – winged helix, H1 – helical 1, H2 – helical 2, FN3 – fibronectin 3) (i) DEAH/RHA proteins have varying N-terminal domains, but show a very high degree of conservation in their C-termini, especially among the spliceosomal DEAH proteins. It is possible that DEAH proteins and perhaps even most DEAH/RHA proteins show a conserved domain organization of their C-termini. Shown is the domain organization of Prp43 [53]. The domain organization of the C-terminus, with the exception of the OB-fold domain, resembles that of Ski2-like proteins [52,95,96]. (WH* - degenerated winged helix, Ratchet corresponds to H1 and H1 in the Ski2-like proteins) (k) NS3/NPH-II proteins have pronounced C- and N-terminal domains, but with the exception of the shown helical C-terminus of NS3 proteins from flaviviridae [51], no further information about these domains is available. (l) Upf1-like proteins feature variable termini. The location of domains 1B and 1C is conserved in the family. Shown is the domain organization of Upf1. (m) Pif1-like proteins feature variable termini, but location of domains 1B is conserved in the family. Domain 2B varies in size from about a dozen residues (RecD1) to more than 100 amino acids (Pif1p). (n) UvrD/Rep proteins also have variable termini. The location of domains 1B and 2B is largely conserved, but domain 2B is absent in LBA1 and HelD. LBA1 also features 3 ankyrin repeats inserted in the helicase domain 1 before domain 1B.