Table 1. Data and final model statistics.
Values in parentheses are for the highest resolution bin.
| 3mwo, new solution | 3ks1, original solution | |
|---|---|---|
| Data statistics | ||
| Space group | P21 | |
| Unit-cell parameters (Å, °) | a = 84.0, b = 41.1, c = 73.6, β = 109.3 | |
| No. of unique reflections | 90672 (8910) | |
| Resolution (Å) | 17.3–1.4 (1.45–1.40) | |
| Rmerge† (%) | 6.4 (19.4) | |
| I/σ(I) | 14.5 (6.1) | |
| Completeness (%) | 96.8 (95.7) | |
| Redundancy | 3.1 (3.1) | |
| Final model statistics | ||
| Rcryst‡ (%) | 16.6 | 20.4 |
| Rfree§ (%) | 18.4 | 23.7 |
| Residue Nos. | 4–261 | 4–261 |
| No. of protein atoms (including alternate conformations) | 4196 | 6177 |
| No. of H2O molecules | 745 | 536 |
| R.m.s.d. for bond lengths (Å) | 0.009 | 0.006 |
| R.m.s.d. for bond angles (°) | 1.3 | 1.1 |
| Ramachandran statistics (%) | ||
| Most favored regions | 87.5 | 88.1 |
| Additionally allowed regions | 12.0 | 11.4 |
| Generously allowed regions | 0.5 | 0.5 |
| B factors (Å2) | ||
| Main chain | 11.0 | 19.9 |
| Side chain | 15.0 | 15.5 |
| Solvent | 27.7 | 19.1 |
R
merge = 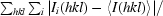
 .
.
R
cryst = 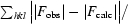
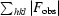 .
.
R free is calculated in the same manner as R cryst, except that it uses 5% of the reflection data that were omitted from refinement.
