Table 1. Likelihood ratio test between the neighbor-dependent and hybrid models of metabolic evolution.
| Pathway map | Neighbor-dependent model | Hybrid model | LH ratio |
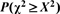
|
||
(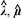 ) ) |
Log LH | (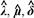 ) ) |
Log LH | |||
| Glycolysis/Gluconeogenesis | (2.6177, 0.4229) | −76.53 | (0.4989, 0.1598, 0.2152) | −63.47 | 26.13 |
|
| Pentose phosphate pathway | (0.5680, 0.7144) | −60.13 | (0.4762, 0.2953, 0.4259) | −53.42 | 13.41 |
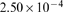
|
| Lysine degradation | (0.0127, 1.0780) | −59.43 | (0.0063, 0.2926, 0.0159) | −52.40 | 14.05 |
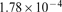
|
| Histidine metabolism | (0.7669, 0.3895) | −54.22 | (0.1852, 0.1643, 0.1370) | −47.28 | 13.89 |
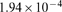
|
| Phenylalanine metabolism | (1.1035, 0.6856) | −62.40 | (1.0299, 1.0297, 0.0038) | −49.91 | 24.97 |
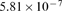
|
| Pyruvate metabolism | (0.1648, 0.5656) | −88.64 | (0.0897, 0.1913, 0.1194) | −81.74 | 13.81 |
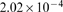
|
The maximum likelihood estimates (MLEs) of the parameter values ( : insertion rate,
: insertion rate,  : deletion rate and
: deletion rate and  : neighbor dependence probability), maximum log-likelihoods, likelihood (LH) ratios, and the
: neighbor dependence probability), maximum log-likelihoods, likelihood (LH) ratios, and the  -values for different pathway maps in P. fluorescens Pf0-1 used in this analysis. The MLEs were obtained using the Gibbs sampler described by Mithani et al.
[9] by evolving the networks from P. fluorescens Pf-5 to P. fluorescens Pf0-1. The equilibrium probability of a network was used as the likelihood of observing the network. The low
-values for different pathway maps in P. fluorescens Pf0-1 used in this analysis. The MLEs were obtained using the Gibbs sampler described by Mithani et al.
[9] by evolving the networks from P. fluorescens Pf-5 to P. fluorescens Pf0-1. The equilibrium probability of a network was used as the likelihood of observing the network. The low  -values for all the pathway maps suggest a better fit for the hybrid model compared to the neighbor-dependent model.
-values for all the pathway maps suggest a better fit for the hybrid model compared to the neighbor-dependent model.
