Table 2. Posterior expectation and variance of evolution parameters estimated using the Gibbs sampler under the hybrid model.
| Pathway Map | Phylogeny |
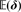
|
var( ) ) |
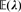
|
var( ) ) |
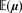
|
var( ) ) |
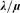
|
| Glycolysis/Gluconeogenesis | ((pfs,pfo),pfl) | 0.2276 | 0.0054 | 2.0552 | 1.0280 | 1.2228 | 0.2653 | 1.6807 |
| (MAP00010) | (pst,(psb,psp)) | 0.2404 | 0.0070 | 1.8645 | 3.3775 | 0.7280 | 0.2891 | 2.5611 |
| 17 pseudomonads | 0.1506 | 0.0034 | 0.7610 | 0.0117 | 0.7329 | 0.0114 | 1.0383 | |
| Pentose phosphate pathway | ((pfs,pfo),pfl) | 0.2785 | 0.0057 | 2.1103 | 0.5582 | 1.7194 | 0.3227 | 1.2273 |
| (MAP00030) | (pst,(psb,psp)) | 0.3251 | 0.0071 | 1.8490 | 1.6921 | 1.0172 | 0.3212 | 1.8178 |
| 17 pseudomonads | 0.1863 | 0.0042 | 0.6762 | 0.0029 | 0.7462 | 0.0126 | 0.9062 | |
| Lysine degradation | ((pfs,pfo),pfl) | 0.0802 | 0.0032 | 0.9662 | 0.2795 | 1.6567 | 1.5943 | 0.5832 |
| (MAP00310) | (pst,(psb,psp)) | 0.0637 | 0.0025 | 0.6986 | 0.1030 | 2.7245 | 2.8663 | 0.2564 |
| 17 pseudomonads | 0.0473 | 0.0030 | 0.4706 | 0.3492 | 0.6443 | 0.7188 | 0.7304 | |
| Histidine metabolism | ((pfs,pfo),pfl) | 0.1833 | 0.0065 | 1.6829 | 1.2133 | 1.0507 | 0.3456 | 1.6017 |
| (MAP00340) | (pst,(psb,psp)) | 0.1749 | 0.0064 | 1.5321 | 0.9479 | 1.0735 | 0.3082 | 1.4272 |
| 17 pseudomonads | 0.0986 | 0.0022 | 0.8685 | 0.0203 | 0.6795 | 0.0057 | 1.2781 | |
| Phenylalanine metabolism | ((pfs,pfo),pfl) | 0.0783 | 0.0029 | 1.1686 | 0.2072 | 1.8255 | 0.9345 | 0.6402 |
| (MAP00360) | (pst,(psb,psp)) | 0.0678 | 0.0024 | 1.0573 | 0.1448 | 2.2334 | 1.1112 | 0.4734 |
| 17 pseudomonads | 0.0617 | 0.0017 | 0.6004 | 0.0061 | 1.0723 | 0.0682 | 0.5599 | |
| Pyruvate metabolism | ((pfs,pfo),pfl) | 0.1413 | 0.0018 | 1.6497 | 0.3362 | 1.7913 | 0.4424 | 0.9210 |
| (MAP00620) | (pst,(psb,psp)) | 0.1559 | 0.0020 | 1.5542 | 0.5376 | 1.2840 | 0.2888 | 1.2105 |
| 17 pseudomonads | 0.1119 | 0.0007 | 0.7668 | 0.0099 | 0.6838 | 0.0142 | 1.1213 |
Posterior expectation and variance of parameter values ( : neighbor dependence probability,
: neighbor dependence probability, 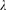 : insertion rate and
: insertion rate and  : deletion rate) estimated using the Gibbs sampler under the hybrid model for the phylogenies relating the bacteria belonging to genus Pseudomonas (Figure 6). Hyperedges common to all seventeen genome-sequenced strains were defined as core and hyperedges missing in all seventeen strains were defined as prohibited hyperedges. The values are averaged over three runs of 60,000 iterations for P. fluorescens and P. syringae phylogenies, and 110,000 iterations for the phylogeny connecting the seventeen Pseudomonas strains with the first 10,000 iterations regarded as burn-in period in each case. Samples were collected every
: deletion rate) estimated using the Gibbs sampler under the hybrid model for the phylogenies relating the bacteria belonging to genus Pseudomonas (Figure 6). Hyperedges common to all seventeen genome-sequenced strains were defined as core and hyperedges missing in all seventeen strains were defined as prohibited hyperedges. The values are averaged over three runs of 60,000 iterations for P. fluorescens and P. syringae phylogenies, and 110,000 iterations for the phylogeny connecting the seventeen Pseudomonas strains with the first 10,000 iterations regarded as burn-in period in each case. Samples were collected every  iteration. The codes MAPxxxxx correspond to the respective KEGG pathway codes [22]. Strain abbreviations: pfl: P. fluorescens Pf-5, pfo: P. fluorescens Pf0-1, pfs: P. fluorescens SBW25, psb: P. syringae pv. syringae B728a, psp: P. syringae pv. phaseolicola 1448A, and pst: P. syringae pv. tomato DC3000.
iteration. The codes MAPxxxxx correspond to the respective KEGG pathway codes [22]. Strain abbreviations: pfl: P. fluorescens Pf-5, pfo: P. fluorescens Pf0-1, pfs: P. fluorescens SBW25, psb: P. syringae pv. syringae B728a, psp: P. syringae pv. phaseolicola 1448A, and pst: P. syringae pv. tomato DC3000.
