Abstract
Protein translation ends when a stop codon in a gene’s messenger RNA transcript enters the ribosomal A site. Mutations that create premature stop codons (nonsense mutations) typically cause premature translation termination. An alternative outcome, read-through translation (or nonsense suppression), is well known in prokaryotic, viral, and yeast genes but has not been clearly documented in humans except in the context of pharmacological manipulations. Here, we identify and characterize native read-through of a nonsense mutation (R201X) in the human copper transport gene, ATP7A. Western blotting, in vitro expression analyses, immunohistochemistry, and yeast complementation assays using cultured fibroblasts from a classical Menkes disease patient all indicated small amounts of native ATP7AR201X read-through and were associated with a dramatic clinical response to early copper treatment.
Introduction
In eukaryotic genes, stop codons, and mutations that produce them, signal proper or premature termination of protein translation, respectively1. Selenoproteins, in which the stop codon UGA encodes selenocysteine2, pose rare exceptions to this rule. Another alternative outcome, read-through translation, is well known in prokaryotic and viral genes3–6, mediated by tRNA mispairing, suppressor tRNAs, or ribosomal jumping but is seldom observed in eukaryotes7–9.
Menkes disease is a X-linked recessive disorder of copper transport caused by diverse mutations in a copper transporting ATPase, ATP7A10. Newborns with this typically fatal neurodegenerative condition may respond well to early copper treatment if their mutation does not completely abrogate ATP7A function11. We previously proposed that internal initiation or translation re-initiation could mediate favorable response to treatment in the context of a premature stop signal in the 5’ region of this locus12, and in vitro evidence for re-initiation was recently reported in association with deletion of ATP7A exons 3 and 413.
In the current study, we evaluated the molecular mechanisms underlying an unexpectedly successful response to early copper treatment in a classical Menkes disease patient with a nonsense mutation in the amino terminal region of ATP7A.
Subjects and Methods
Case Report
The patient was born at 37½ weeks gestation by normal spontaneous vaginal delivery with a birth weight of 3.1 kg. A cephalohematoma was noted over the left parieto-occipital scalp. There was a slight amount of hair present on the sides of the head and the hair was light in color with a fine texture. Low temperature, once on the first afternoon of life, and hyperbilirunemia (peak blood concentration 15.5 mg/dl) were noted and resolved. The family history was notable for a prior sibling affected with Menkes disease who was profoundly neurologically delayed at the time of his death at 6 months of age following a sudden and massive gastrointestinal hemorrhage14. Given that recurrence risk for subsequent male offspring was potentially as high as 50%, the subject of this report underwent careful biochemical evaluation in the immediate postnatal period and was diagnosed has having Menkes disease in the first week of life based on elevated placental copper level (J. Centeno, Ph.D., Armed Forces Institute of Pathology, Washington, DC), and abnormal plasma and cerebrospinal fluid catecholamine levels (C. Holmes, D. Goldstein, NIH). The infant began copper injections under an Institutional Review Board-approved protocol (ClinicalTrials.gov identifier NCT00001262), at eight days of age, with the informed consent of his parents. The study drug was copper histidine, FDA Investigational New Drug (#34,166) holder: SG Kaler, prepared by the NIH Pharmaceutical Development Service, as previously described15. The dose regimen was 250 µg by subcutaneous injection twice daily to age 1 year, and 250 µg once daily thereafter to age 3 years when it was stopped, as indicated by the protocol.
Tissue Culture
R201X and normal fibroblast cultures were established from explant cultures of full-thickness, skin punch biopsies. Cells were grown in MEM-Alpha medium (InVitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS, Hyclone, Logan, UT) and 2 mM l-glutamine, and were used before passage 10.
Mutation Analysis and DNA sequencing
Mutation analysis was performed as previously described16.
Antibody Production
Polyclonal antisera against ATP7A was obtained by immunizing rabbits with peptides containing amino acid residues 146–160 (N-terminal antibody) and 1483–1500 (C-terminal antibody).
Western blot analysis
Fibroblast microsomal protein fractions were obtained by ultracentrifugation (10,000g × 60m) following cell lysis and Western analyses were performed as previously described17. 100 µg microsomal protein was loaded in each lane.
Densitometric Quantitation
Western blots were scanned to create digital grey-scale images, and imported into ImageJ (http://rsbweb.nih.gov/ij/download.html) for densitometric analysis, as previously described17.
Confocal Microscopy
Immunohistochemical analyses were performed as previously described11.
Yeast complementation
Functional copper transport associated with normal, R201X, and Q1385X ATP7A alleles was assessed in a yeast complementation assay, as previously described11,17,18.
Timed Growth Assay
A timed yeast growth assay to estimate the percentage of copper transport by the R201X mutant allele relative to normal was performed as previously described18.
In vitro Expression
Reporter genes were constructed to mimic the in vivo molecular context, generating peptides 249 amino acids in length that spanned the region of the nonsense mutation. These cDNAs were cloned into pTR-UF11 to express them from the CMV enhancer/chicken ß-actin promoter. Reporter constructs were transfected into HEK-293 cells by standard calcium phosphate transfection. Cells were harvested 72 hours post-transfection and mechanically homogenized in CelLytic M Cell Lysis Reagent (Sigma). Western blots were performed with an ATP7A N-terminal antibody.
Results
Clinical Outcome
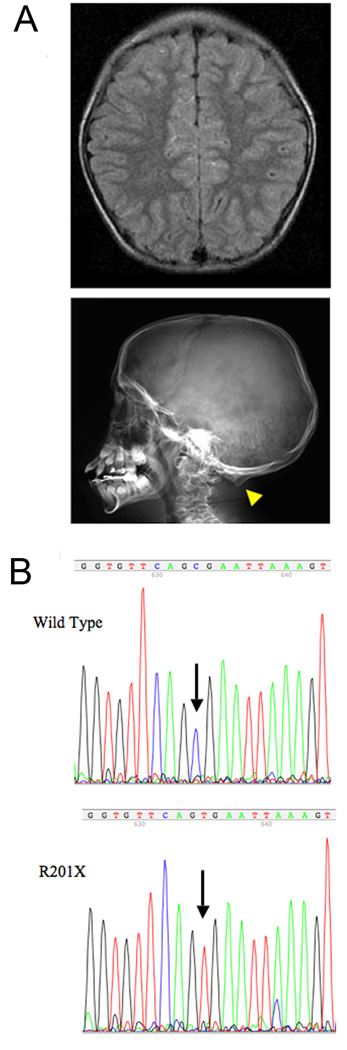
At 2, 8 and 13 months of age, the patient’s serum copper levels were 79, 129, and 146 µg/dl (normal range 70–150 µg/dl), respectively. He had no seizures or electroencephalographic abnormalities, and walked independently at 16 months of age. Brain magnetic resonance imaging at 2 and 3 years of age showed appropriate myelination and no cerebral or cerebellar atrophy (Fig. 1a, top panel). Copper injection treatment was discontinued at 3 years of age, as called for by the clinical trial. On follow-up evaluation at 9 ½ years of age, the patient was a 4th grade student earning above average grades in regular (not special needs) classes and had no physical limitations. He was an intelligent conversationalist, and his neurological examination was non-focal. When seen at 11½ years of age, he continued to appear healthy and manifested no cognitive or neurological problems. Skull radiographs disclosed occipital exostoses (Fig. 1a, bottom panel), a phenotypic hallmark in long-surviving Menkes disease patients19.
Figure 1. Molecular and Radiologic findings in Patient with Menkes disease.
A. Upper panel: Axial flair magnetic resonance image of the patient’s brain at 3 years of age shows normal volume and myelination (hypointense signal). (b) Lateral skull radiograph at age 11.5 years shows occipital exostoses (arrowhead). B. Sequence of cDNA illustrating a cytosine (C) to thymine (T) transition producing the R201X mutation in ATP7A.
Mutation Analysis and Characterization
We identified a novel C to T transition at nucleotide 746 in exon 3 of the ATP7A coding sequence, changing codon 201 from CGA to UGA (Fig. 1b) in two affected members of a classical Menkes disease family. In the context of an excellent neurologic outcome in the younger member whom we diagnosed at birth and treated with copper injections for the first three years of life, we suspected that translation re-initiation downstream of R201X produced a small quantity of a truncated but partially functional copper transporter.
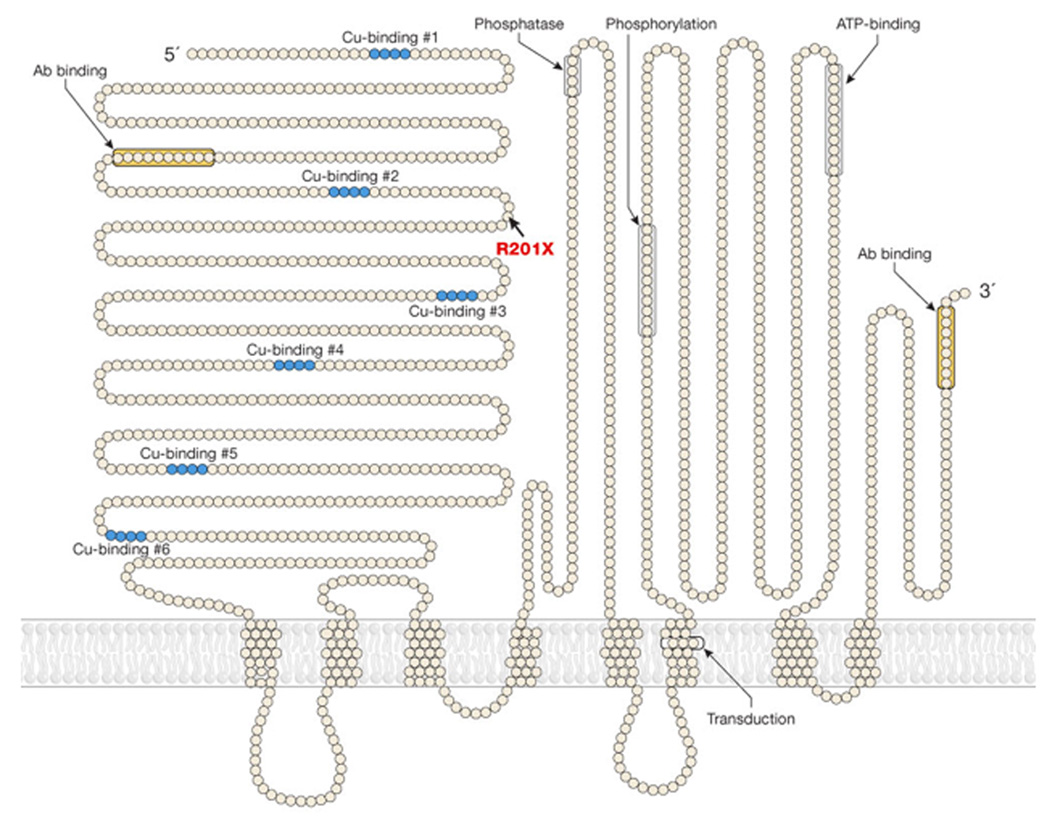
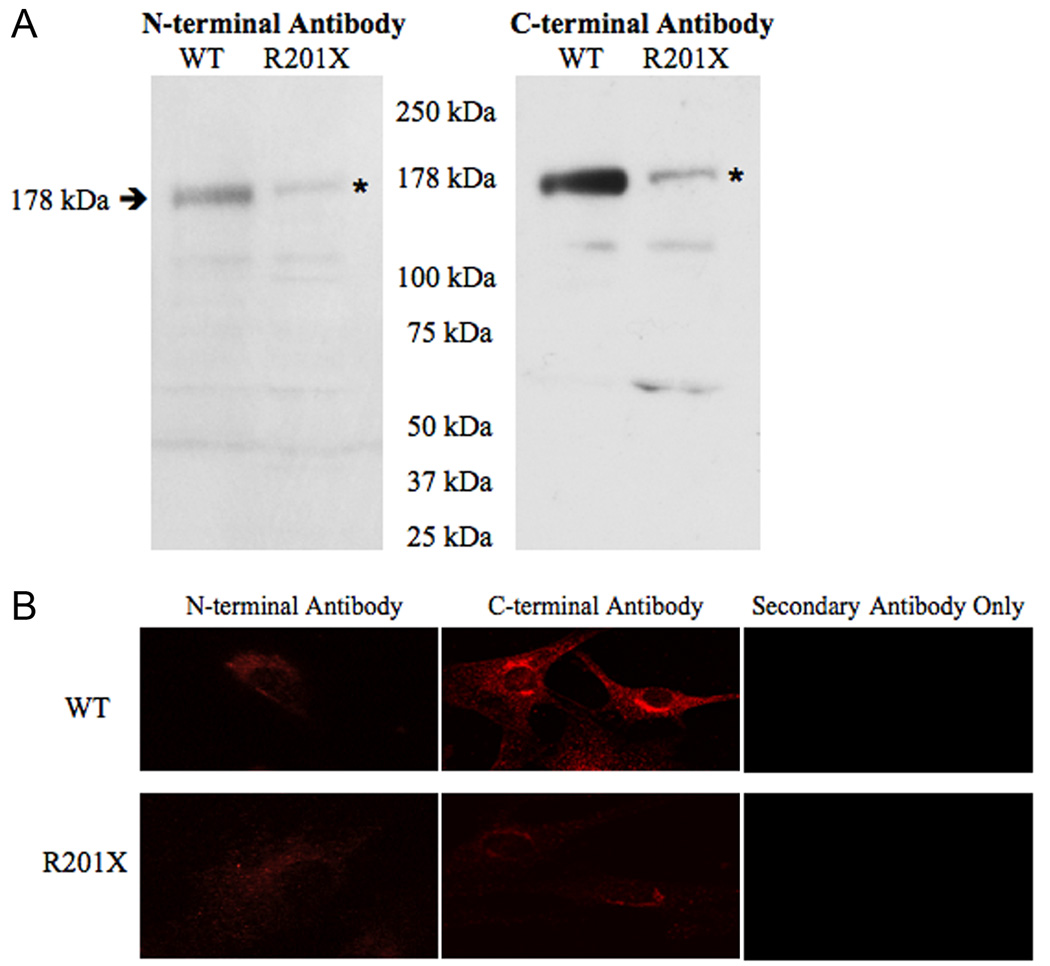
We performed Western analyses of microsomal protein extracts from R201X patient fibroblasts using antibodies generated against amino- and carboxyl-terminal segments of human ATP7A (see model for antibody location, Fig. 2). We predicted detection of truncated forms with the latter anti-sera. Unexpectedly however, both amino- and carboxyl-terminal antibodies detected small amounts of the full-length (178 kDa) ATP7A protein in the R201X fibroblasts, and no products of size consistent with re-initiation closely downstream of R201X (Fig. 3a). Re-initiation at downstream AUG codons predicts that such smaller products would be detectable only with the C-terminal antibody, and the sole possible candidate on Western blots was approximately 64 kDa in size (Fig. 3a, right panel, right lane) which would lack many of the critical functional domains of the normal 178 kDa ATP7A (Fig. 2). While these results render downstream re-initiation unlikely, our findings are consistent with translational read-through of R201X through which small quantities of full length ATP7A detectable with either antibody are generated. Reverse transcription-polymerase chain reaction of the relevant ATP7A segment using total RNA from R201X fibroblasts, and sequencing confirmed the C to T transition in cDNA, excluding RNA editing as an explanation for the presence of full length protein in these cells. Densitometric quantitation of the two Westerns, corrected for loading, indicated that R201X cells produced 5.7% (detected by the N-terminal antibody) and 6.4% (detected by the C-terminal antibody) of the wild type amount of full length ATP7A.
Figure 2. Model of the ATP7A Copper Transporter.
The R201X mutation is located between the second and third copper-binding motifs. The amino- and carboxy-terminal antibody binding locations are noted (orange color).
Figure 3. Evidence of Translational Read-through of R201X in ATP7A.
A. Western blots of fibroblast microsomal protein from a normal (WT) control male and Menkes disease patient with the R201X mutation show that the full length (178 kDa) gene product is detected with an amino-terminal antibody generated against amino acids 146–160 of ATP7A (left hand panel), as well as with a carboxy-terminal antibody against residues 1479–1500 of ATP7A (right hand panel). The quantity is reduced in the R201X cells (note asterisks). The C-terminal antibody gives a stronger signal than the N-terminal antibody. B. Confocal microscopy of fibroblasts from normal (WT) control (top panels) and R201X patient (bottom panels) stained with N-terminal and C-terminal antibodies to ATP7A, and with Texas-Red labeled anti-rabbit IgG secondary antibody alone, as a negative control. The normal cells show perinuclear staining consistent with trans-Golgi localization, a pattern also seen in R201X cells at much lower intensity. The C-terminal antibody gives a stronger signal than the N-terminal antibody.
Immunohistochemical studies also detected trace amounts of an anti-ATP7A reacting material in R201X cells (Fig. 3b, lower panels). The perinuclear staining pattern in these cells was comparable in quality to that in wild type fibroblasts, and consistent with the known trans-Golgi localization of ATP7A20.
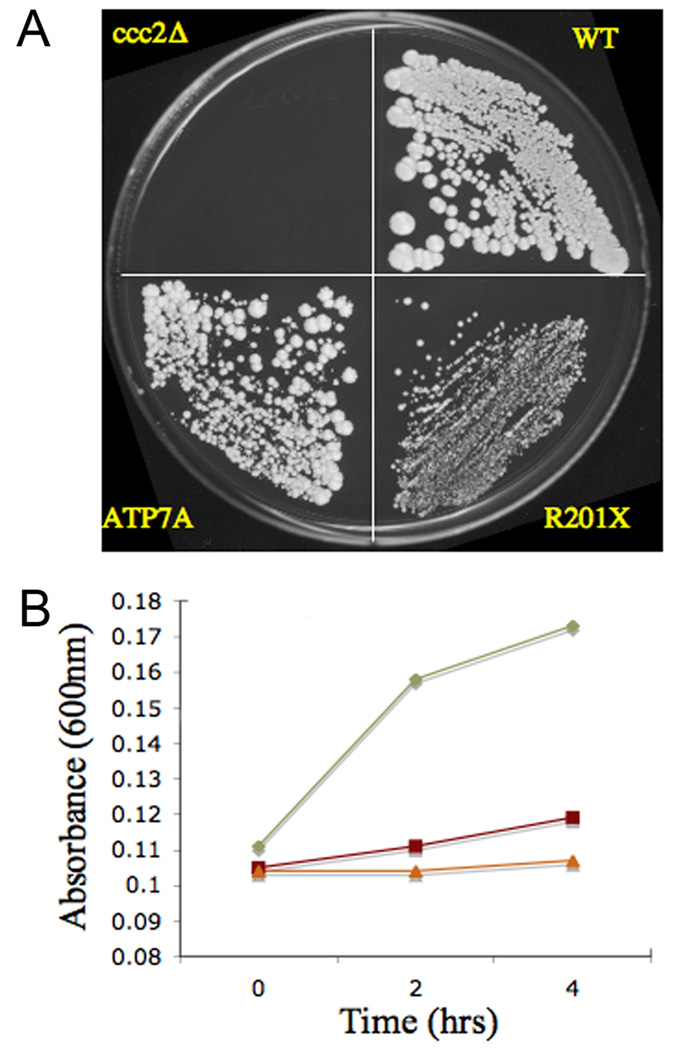
Next, we assessed whether functional copper transport was associated with the R201X ATP7A allele, utilizing a yeast complementation assay (Fig. 4a). The S. cerevisiae strain ccc2Δ is unable to grow on nutritionally restricted (low copper and low iron) media, however transformation with wild type ATP7A cDNA restores normal growth. Transformation with R201X cDNA complemented ccc2Δ sufficiently for partial restoration of growth in both solid and liquid media. Using a timed growth assay that we described previously15, we calculated the estimated residual copper transport capacity of the R201X allele to be 5–10% of wild type (Fig. 4b).
Figure 4. Yeast Complementation Documents Functional Copper Transport Associated with R201X.
A. On copper/iron-deficient media, the copper transport mutant ccc2Δ fails to grow, whereas the WT strain grows normally. Transformation of ccc2Δ with the R201X mutant allele restores growth although less robustly than when transformed with the wild type ATP7A. B. Timed growth assays18 estimate R201X growth in the range of 5–10% compared to wild type (WT). Diamond = wild type, Square = R201X, Triangle = ccc2Δ.
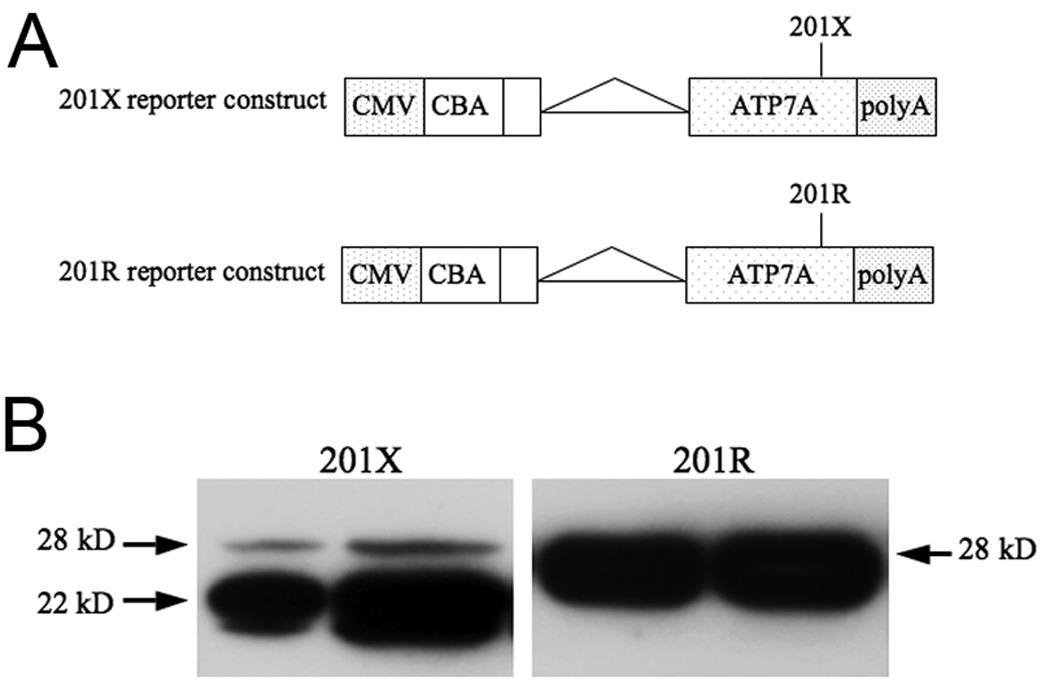
Finally, we performed in vitro expression experiments to express the initial 249 amino acids of ATP7A, using cDNA constructs containing either the wild type or mutant sequence for residue 201 in HEK-293 cells by transfection with the expression vector pTR-UF11 (Fig. 5a). Western analysis showed the predicted 28kD peptides in both, although the amount was much lower in the mutant (Fig. 5b, left panel), as expected. The mutant construct also showed a prominent 22 kDa species, corresponding to the truncated peptide produced when the R201X nonsense codon was not suppressed.
Figure 5. In Vitro Expression Analyses of 201X Indicate Full-length and Truncated ATP7A Peptides.
A. Reporter constructs for 201X and 201R in the expression vector pTR-UF11. CMV = cytomegalovirus enhancer; CBA = chicken ß-actin promoter; ATP7A = cDNA sequence encoding the initial 249 amino acids of ATP7A; polyA = polyadenylation sequence. B. Western blots of HEK-293 protein following transient expression detect the full length peptide (28 kDa) using a N-terminal ATP7A antibody with both constructs, as well as a smaller form (22 kDa) corresponding in size to the prematurely terminated peptide (200 amino acids) from the mutant construct.
Discussion
Our patient’s excellent treatment response, in conjunction with detection of full length ATP7A in R201X fibroblasts by Western blotting, discernible immunofluorescence on confocal microscopy of R201 cells, functional copper transport by the mutant allele in a yeast complementation assay, and the in vitro expression results all support the conclusion of ATP7AR201X read-through. Since Menkes disease is a X-linked recessive disorder, partial expression of the full length gene product by a second mutant allele (compound heterozygosity) is not possible and does not explain the finding.
In mammalian selenoproteins, such as glutathione peroxidase and iodothyronine 5’ deiodinase, UGA encodes selenocysteine instead of causing translation termination2. Since S. cerevisiae does not contain this amino acid, the yeast complementation results presented here suggest that an alternative mechanism is active. In the rabbit β-globin gene, variable ribosomal hopping and tRNA mispairing are associated with normal stop codon read-through9. Amino acid sequencing of the 28 kDa peptide expressed by the R201X reporter construct will be required to clarify these or other mechanisms as the precise means of ATP7AR201X read-through. Our experiments suggest that the amount of nonsense suppression is not high, only about 6% compared to normal, and we were unable to isolate the R201X read-through product in a quantity and purity sufficient for this purpose.
Table 1A summarizes examples of UGA nonsense codon read-through in eukaryotes, with emphasis on the associated sequence contexts7–9. The ATP7AR201X context resembles that for native read-through in the yeast genes STE6 and ADE2, and in rabbit beta-globin, by possessing a cytosine/adenine dinucleotide upstream of the stop codon (−3 and −2 positions). For ADE2, this sequence was associated with read-through efficiency ranging from 10–16%8. Our findings lend support to the concept that unique 5’ sequences have a critical role in nonsense suppression and that mRNA structure, through interaction with the ribosome, may modulate competition between eukaryotic release factors and suppressor tRNAs. We reviewed the 5’ sequence context for eight ATP7A nonsense mutations that we identified in unrelated Menkes disease patients, and none shared the R201X sequence context (Table 1B).
Table 1.
Sequence Contexts Associated with Eukaryotic UGA Read-through
| A. Sequence Contexts Associated with Eukaryotic UGA Read-through | ||
|---|---|---|
| Gene | Organism | Sequence |
| STE67 | S. cerevisiae | GGG CAA UGA CAA AGA |
| ADE28 | S. cerevisiae | ACC CAA UGA CAG UUA |
| Beta globin9 | Oryctolagus cuniculus | UAC CAC UGA GAU CUU |
| ATP7AR201X | H. sapiens | GUU CAG UGA AUU AAA |
| B. Sequence Contexts Associated with Other Nonsense Mutations in ATP7A | ||
| Gene | Mutation | Sequence |
| ATP7A | K155X | CUG GAG UAA AAG UCA |
| Q168X | AUG GCU UAA GCU GGU | |
| R409X | UCC AUA UGA GUC UCC | |
| Y594X | AUC CUA UAG UGC UCC | |
| R645X | CAU AAA UGA GAA AUA | |
| R795X | CUA GGC UGA UGG CUG | |
| R980X | AUC UCC UGA ACA GAA | |
| Q1385X | GUU UUG UAG CCC UGG | |
All stop codons are shown in bold.
Conforming bases in the read-through sequence contexts are shown in red.
Regardless of the precise mechanism, read-through of ATP7AR201X in this instance was directly relevant to a remarkable treatment response in a patient with Menkes disease, a neurogenetic disorder that is often lethal. Early diagnosis and treatment with copper injections, when associated with missense or splicing mutations that retain some copper transport capacity, may greatly improve clinical outcomes in this disease11. While the successful clinical outcome reported here involved an unusual example of native translational read-through, it highlights the distinct possibility that aminoglycosides or alternative agents that promote read-through21–24 may be useful approaches for other Menkes disease patients with nonsense mutations.
Acknowledgements
We thank the patient and his family for their participation in the clinical trial, the staffs of the NICHD Microscopy and Imaging Core, the NINDS DNA Sequencing Facility, and the NIH Clinical Center for support of these investigations, and Richard Maraia and Dolph Hatfield for thoughtful review of the manuscript. This work is supported by the NIH Intramural Research Program.
Footnotes
Author Information: The authors declare no competing financial interests.
References
- 1.Amrani N, Sachs MS, Jacobson A. Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol. 2006;7:415–425. doi: 10.1038/nrm1942. [DOI] [PubMed] [Google Scholar]
- 2.Berry MJ, Banu L, Chen YY, Mandel SJ, Kieffer JD, Harney JW, Larsen PR. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3' untranslated region. Nature. 1991;353:273–276. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- 3.Schecter JB. Suppressed nonsense mutations in the araC gene of Escherichia coli provide three novel variant proteins. J Bacteriol. 1983;154:1329–1338. doi: 10.1128/jb.154.3.1329-1338.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baranov PV, Fayet O, Hendrix RW, Atkins JF. Recoding in bacteriophages and bacterial IS elements. Trends Genet. 2006;22:174–181. doi: 10.1016/j.tig.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Skuzeski JM, Nichols LM, Gesteland RF, Atkins JF. The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J Mol Biol. 1991;218:365–373. doi: 10.1016/0022-2836(91)90718-l. [DOI] [PubMed] [Google Scholar]
- 6.Beier H, Grimm M. Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res. 2001;29:4767–4782. doi: 10.1093/nar/29.23.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fearon K, McClendon V, Bonetti B, Bedwell DM. Premature translation termination mutations are efficiently suppressed in a highly conserved region of yeast Ste6p, a member of the ATP-binding cassette (ABC) transporter family. J Biol Chem. 1994;269:17802–17808. [PubMed] [Google Scholar]
- 8.Tork S, Hatin I, Rousset JP, Fabret C. The major 5' determinant in stop codon read-through involves two adjacent adenines. Nucleic Acids Res. 2004;32:415–421. doi: 10.1093/nar/gkh201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chittum HS, Lane WS, Carlson BA, Roller PP, Lung FD, Lee BJ, Hatfield DL. Rabbit β-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry. 1998;37:10866–10870. doi: 10.1021/bi981042r. [DOI] [PubMed] [Google Scholar]
- 10.Kaler SG. Barness LA. Advances in Pediatrics. Volume 41. St. Louis: CV Mosby; 1994. Menkes disease; pp. 263–304. [PubMed] [Google Scholar]
- 11.Kaler SG, Holmes CS, Goldstein DS, Tang JR, Godwin SC, Donsante A, Liew CJ, Sato S, Patronas N. Neonatal Diagnosis and Treatment of Menkes Disease. N Engl J Med. 2008;358:605–614. doi: 10.1056/NEJMoa070613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaler SG. Menkes disease mutations and response to early copper histidine treatment. Nature Genet. 1996;13:21–22. doi: 10.1038/ng0596-21. [DOI] [PubMed] [Google Scholar]
- 13.Paulsen M, Lund C, Akram Z, Winther JR, Horn N, Moller LB. Evidence that translation reinitiation leads to a partially functional Menkes protein containing two copper-binding sites. Am J Hum Genet. 2006;79:214–229. doi: 10.1086/505407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaler SG, Westman JA, Bernes SM, Elsayed AM, Bowe CM, Freeman KLB, Wu CD, Wallach MT. Gastrointestinal hemorrhage associated with gastric polyps in Menkes disease. J Pediatr. 1993;122:93–95. doi: 10.1016/s0022-3476(05)83496-1. [DOI] [PubMed] [Google Scholar]
- 15.Kaler SG, Das S, Levinson B, Goldstein DS, Holmes CS, Patronas NJ, Packman S, Gahl WA. Successful early copper therapy in Menkes disease associated with a mutant transcript containing a small in-frame deletion. Biochem Mol Med. 1996;57:37–46. doi: 10.1006/bmme.1996.0007. [DOI] [PubMed] [Google Scholar]
- 16.Liu P-C, McAndrew PE, Kaler SG. Rapid and robust screening of the Menkes disease/occipital horn syndrome gene. Genet Test. 2002;6:255–260. doi: 10.1089/10906570260471778. [DOI] [PubMed] [Google Scholar]
- 17.Donsante A, Tang JR, Godwin SC, Holmes CS, Goldstein DS, Bassuk A, Kaler SG. Differences in ATP7A gene expression underlie intra-familial variability in Menkes disease/occipital horn syndrome. J Med Genet. 2007;44:492–497. doi: 10.1136/jmg.2007.050013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang J, Robertson SP, Lem KE, Godwin SC, Kaler SG. Functional copper transport explains neurologic sparing in occipital horn syndrome. Genet Med. 2006;8:711–718. doi: 10.1097/01.gim.0000245578.94312.1e. [DOI] [PubMed] [Google Scholar]
- 19.Kaler SG, Gallo LK, Proud VK, Percy AK, Mark Y, Segal NA, Goldstein DS, Holmes CS, Gahl WA. Occipital horn syndrome and a mild Menkes phenotype associated with splice site mutations at the MNK locus. Nature Genetics. 1994;8:195–202. doi: 10.1038/ng1094-195. [DOI] [PubMed] [Google Scholar]
- 20.La Fontaine S, Firth SD, Lockhart PJ, Brooks H, Parton RG, Camakaris J, Mercer JF. Functional analysis and intracellular localization of the human menkes protein (MNK) stably expressed from a cDNA construct in Chinese hamster ovary cells (CHO-K1) Hum Mol Genet. 1998;7:1293–1300. doi: 10.1093/hmg/7.8.1293. [DOI] [PubMed] [Google Scholar]
- 21.Howard MT, Anderson CB, Fass U, Khatri S, Gesteland RF, Atkins JF, Flanigan KM. Readthrough of dystrophin stop codon mutations induced by aminoglycosides. Ann Neurol. 2004;55:422–426. doi: 10.1002/ana.20052. [DOI] [PubMed] [Google Scholar]
- 22.Pinotti M, Rizzotto L, Pinton P, Ferraresi P, Chuansumrit A, Charoenkwan P, Marchetti G, Rizzuto R, Mariani G, Bernardi F International Factor VII Deficiency Study Group. Intracellular readthrough of nonsense mutations by aminoglycosides in coagulation factor VII. J Thromb Haemost. 2006;4:1308–1314. doi: 10.1111/j.1538-7836.2006.01915.x. [DOI] [PubMed] [Google Scholar]
- 23.Sermet-Gaudelus I, Renouil M, Fajac A, Bidou L, Parbaille B, Pierrot S, Davy N, Bismuth E, Reinert P, Lenoir G, Lesure JF, Rousset JP, Edelman A. In vitro prediction of stop-codon suppression by intravenous gentamicin in patients with cystic fibrosis: a pilot study. BMC Med. 2007;5:5. doi: 10.1186/1741-7015-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keeling KM, Du M, Bedwell DM. Therapies of nonsense-associated diseases. In: Maquat LE, editor. Nonsense-Mediated mRNA Decay. Georgetown, TX: Landes Bioscience/Eurekah.com; 2006. pp. 121–136. [Google Scholar]