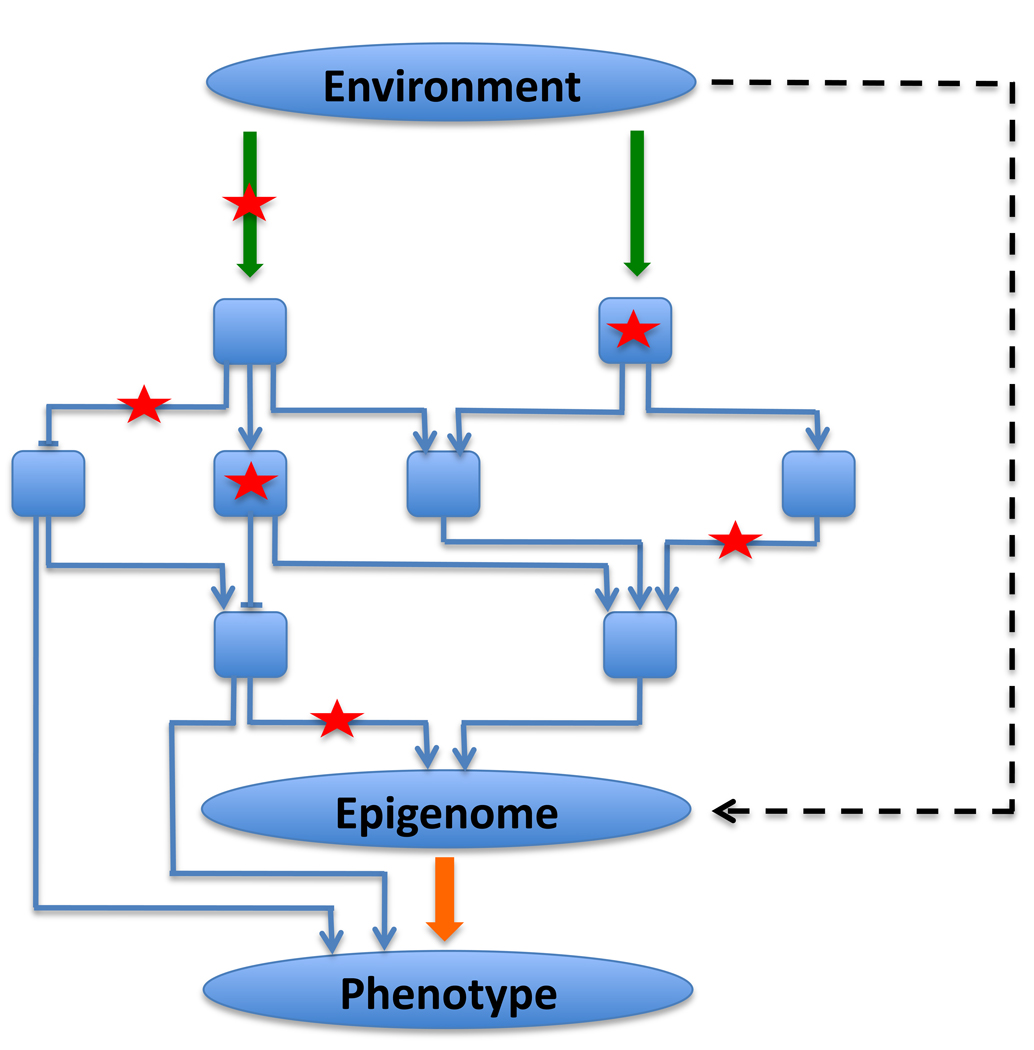
Figure 3. Integrating epigenome information in studies of disease.
An epigenome can be thought of as a sensor of perturbations in the environment, in signal transduction, and in the gene regulatory networks within cells. DNA sequence variations in various components of these systems are shown as red stars. Sequence variations in genes coding for proteins and enzymes of signal transduction pathways (green arrows) and in genes coding for transcription factors (blue boxes) can modulate signaling output from these systems. Sequence variants in cis-regulatory elements (blue lines) can also modulate the signaling output from gene regulatory networks. Although individually subtle and weak, these sequence variants may have a large cumulative combinatorial effect on the epigenome. Thus, an epigenome may integrate epistatic interactions of many genetic loci. Perturbations in environment can affect epigenomes directly (black dotted arrow) or through signaling and gene regulatory cascades that impinge on the chromatin states. Phenotype/disease status of an organism is an outcome of complex interactions between genotype and environment. An epigenome is an intermediate phenotype that is causally ‘closer’ to environmental and genetic factors and thus can provide very useful information for studies attempting to identify disease susceptibility factors. In addition, environmental signals can regulate gene expression levels through post-translational modifications of proteins involved in cell signaling and gene regulation without impacting DNA sequence or epigenetic marks, bypassing the epigenome to directly influence gene expression and phenotype.