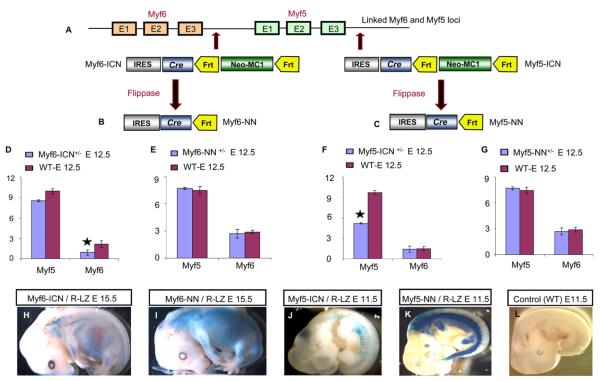
Figure 1. Alleles of myf5 and myf6 Cre Drivers.
(A) myf5 and myf6 genes are linked on chromosome 10. The 3′UTR of myf6 (in myf6-ICN)or myf5 (in myf5-ICN) harbors an encephalomyocarditis Internal Ribosomal Entry Site (IRES) that is linked to the Cre coding sequence and is followed by the Neomycin (neo)-resistance selection cassette with MC1 promoter. The selection cassette with MC1 promoter is flanked by two FRT sequences.
(B and C) Breeding with FLP-e mice leads to recombination between FRT sites, generating the “NN” alleles.
(D–G) Semiquantitative RT-PCR on total RNA from E12.5 embryos shows (D) reduced expression of myf6 in myf6-ICN+/− mice (star) but (E) normal expression in myf6-NN+/− mice compared to wild-type. Similarly, myf5 expression is (F) reduced in myf5-ICN+/− mice (star) but (G) normal in myf5-NN+/− mice compared to wild-type.
(H and I) β-gal staining revealed a reduced myf6 lineage in E15.5 (H) myf6-ICN/R-LZ embryos compared to (I) myf6-NN/R-LZ embryos.
(J and K) Similarly, the myf5 lineage in E11.5 (J) myf5-ICN/R-LZ embryos is reduced compared to (K) myf5-NN/R-LZ embryos.
(L) A control wild-type E11.5 embryo shows an absence of any β-gal staining.