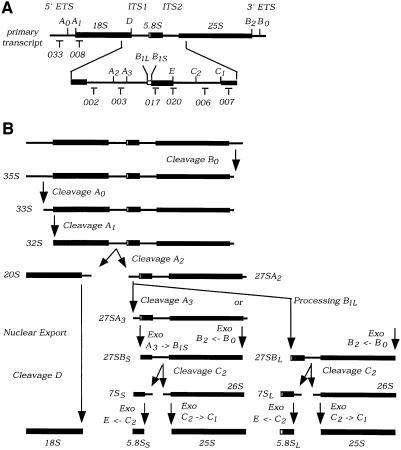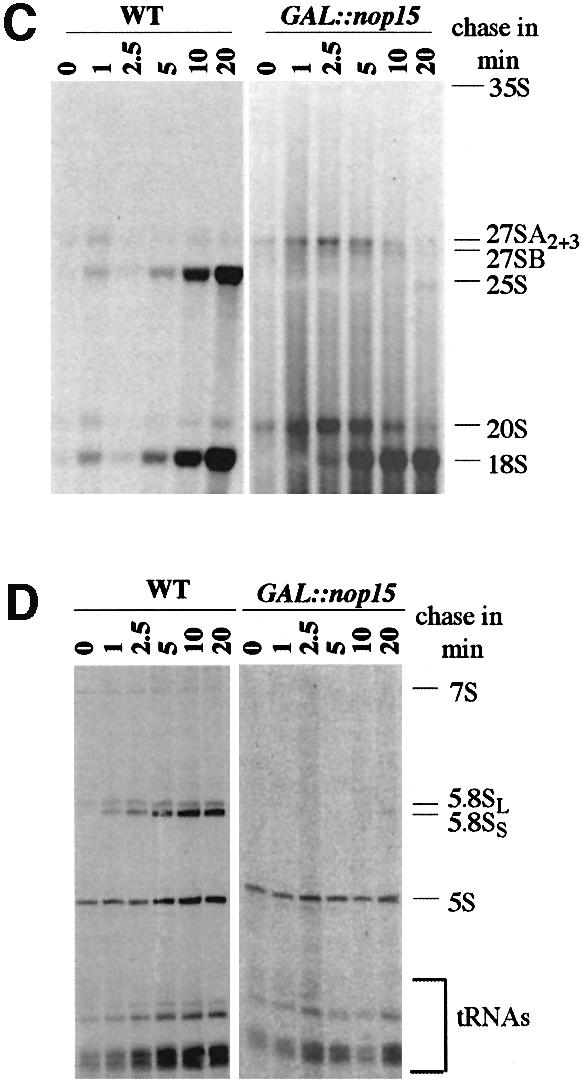
Fig. 2. Pulse–chase analysis of rRNA synthesis. (A) Structure and processing sites of the 35S pre-rRNA. This precursor contains the sequences for the mature 18S, 5.8S and 25S, which are separated by the two internal transcribed spacers ITS1 and ITS2 and flanked by the two external transcribed spacers 5′ETS and 3′ETS. The positions of oligonucleotide probes utilized in northern hybridization and primer extension analyses are indicated. (B) Pre-rRNA processing pathway. For further discussion of the processing pathway and enzymes, see Kressler et al. (1999), Venema and Tollervey (1999), Warner (2001) and Fatica and Tollervey (2002). (C and D) Pre-rRNA was pulse-labelled with [3H]uracil for 2 min at 30°C and chased with a large excess of unlabelled uracil for the times indicated. Labelling was performed for the GAL::nop15 strain and a wild-type strain 16 h after transfer to glucose medium. (C) High molecular weight RNA separated on a 1.2% agarose/formaldehyde gel. (D) Low molecular weight RNA separated on an 8% polyacrylamide/urea gel.