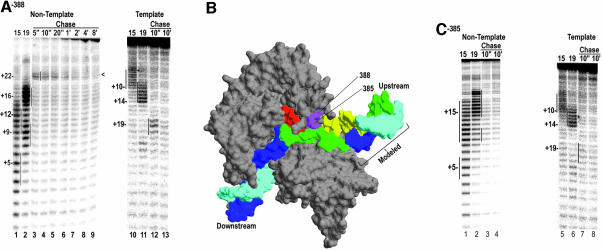
Fig. 2. (A) Non-template (lanes 1–9) or template (lanes 10–13) strand cleavage by Fe-BABE conjugated to aa 388 in ECs halted at +15 (lanes 1 and 10), or +19 (lanes 2 and 11), or chased from +14 with UTP+CTP for the indicated times. Cleavage sites were mapped by reference to G+A ladders prepared with the identical DNAs and are highlighted by vertical lines. (B) Structure of the T7RNAP EC (Yin and Steitz, 2002; pdb:1msw) with the template strand in cyan, the non-template strand in blue, and the RNA in red. Residues 388 and 385 are in magenta and regions of the template and non-template strands cut by the 388 conjugate (in the +15 and +19 halted complexes) are colored yellow and green, respectively. Note that the DNA labeled as ‘Modeled’ was added onto the crystal structure by extending the upstream DNA. (C) As in (A), but for complexes with the conjugate at residue 385.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.