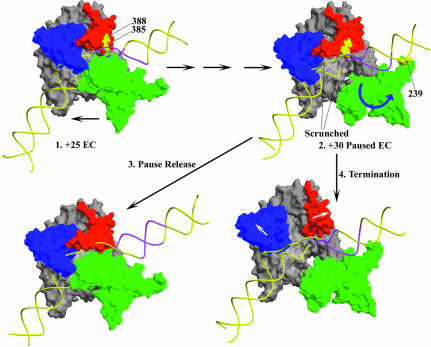
Fig. 8. A model for class II termination. 1. EC crystal structure (pdb: 1HW38; duplex DNA extensions are modeled onto the DNA from the crystal structure and the RNA is omitted for clarity). Polymerase movement is in the direction of the lower arrow. The thumb (aa 330–410, red), fingers (aa 560–680, blue) and N-terminal (aa 60–255, green) domains form a box around the DNA which, barring a conformational change, effectively precludes template release. If the EC is at +25 on the template used in these studies, then the region highlighted in magenta corresponds to the conserved class II element at +16 to +23. 2. Hypothetical +30 Paused EC structure. Interactions between 385/388 and the class II element, first established in the +25 EC, may persist as the RNA is extended to +30. The additional DNA is accommodated in the EC by a combination of ‘scrunching’ and/or conformational changes in the polymerase, and, after reaching +30, the polymerase backtracks ∼3 nt to form the stably paused complex. The N-terminal domain may shift and rotate as suggested by the blue arrow, so that an Fe-BABE conjugate at 239 would cleave upstream of the class II element. This movement of the N-terminal domain could also create room to allow the trailing edge of the transcription bubble to become duplex, as has been observed during pausing at the class II site (Song and Kang, 2001). 3. Spontaneous release of the class II element leads to escape from the pause and recovery of the normal EC structure (now shown at +31). 4. Strain in the paused complex may cause the thumb and fingers to move apart as suggested by the white arrows, so the RNAP can release DNA and terminate transcription.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.