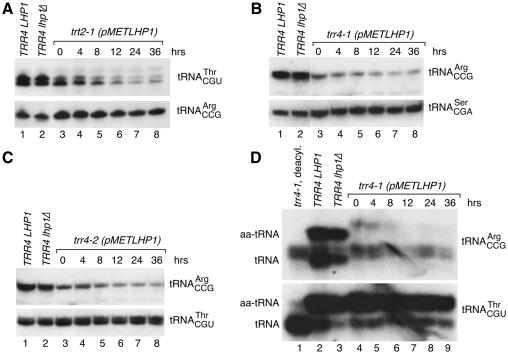
Fig. 2. Depletion of LHP1 from trr4 and trt2 cells. trt2-1 (A), trr4-1 (B) and trr4-2 cells (C) containing pMETLHP1 were grown at 25°C in media lacking methionine and switched to 2 mM methionine media at time 0. At intervals (lanes 3–8), RNA was subjected to northern analysis to detect tRNAThrCGU (A, top; C, bottom), tRNAArgCCG (A, bottom; B and C, top) or tRNASerCGA (B, bottom). RNA was also analyzed from wild-type (lanes 1) and lhp1Δ cells (lanes 2). For unknown reasons, tRNAThrCGU runs as a doublet. (D) At intervals after the switch to 2 mM methionine, RNA was extracted from trr4-1 cells under acidic conditions and fractionated in acidic acrylamide gels. The northern blot was probed to detect tRNAArgCCG and tRNAThrCGU. Lane 1, deacylated trr4-1 RNA. RNA was also analyzed from wild-type cells (lane 2) and cells lacking LHP1 (lane 3). Consistent with an altered structure, charged and uncharged forms of wild-type tRNAArgCCG migrate differently than these forms of trr4-1 tRNAArgCCG. Lane 3 is underloaded.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.