Rhodopsin is a vertebrate dim-light photoreceptor consisting of a seven-helical transmembrane opsin protein and a chromophore, 11-cis retinal, linked to the protein via a protonated Schiff base (PSB).1 Following photon absorption, 11-cis retinal isomerizes to the all-trans configuration to initiate the visual transduction cascade. Since optimal transduction depends on the thermal stability of rhodopsin, the thermal decay process has long been a subject of interest, yet the chemistry behind it has remained unclear.2–4
Baylor et al. demonstrated that the thermal isomerization of rhodopsin, which enhances dark noise and reduces photosensitivity, generates the same physiological response as photoisomerization.5,6 Furthermore, rhodopsin was reported to undergo thermal decay characterized by a decrease in visible absorption at 500 nm and an increase in UV absorption at 380 nm upon incubation at 37–55°C.7,8 The observed thermal decay was attributed to hydrolysis of the PSB. Further analysis by HPLC showed that the concentration of all-trans retinal extracted from rhodopsin increased after incubation in the dark at 55°C. In this case, the thermally-induced isomerization from 11-cis to all-trans was proposed to be initiated by a different process – thermal denaturation.9
We recently proposed that a hydrogen-bonding network at the retinal binding site10 is involved in the counterion switch between dark state rhodopsin and metarhodopsin I, an intermediate precursor to transducin photoactivation.11,12 This theory has prompted our current investigation of the thermal properties of the putative hydrogen-bonding network through solvent deuterium isotope effects. In this report, we attempt to clarify and characterize the chemical reactions responsible for the thermal decay process, with particular emphasis on (1) thermal decay marked by a decrease in optical density at 500 nm (OD500), (2) thermal isomerization of 11-cis to all-trans retinal, and (3) hydrolysis of the PSB. Our results suggest that although PSB hydrolysis and retinal isomerization affect the thermal decay of rhodopsin, denaturation of the secondary structure is not significant. Moreover, we discover that the rates of thermal decay, isomerization, and PSB hydrolysis are two- to three-fold slower in D2O than H2O. Based on these results, we conclude that the rate-determining step of these thermal processes involves the breaking of hydrogen bonds, which probably stabilize the tertiary structure of rhodopsin, thereby contributing to the high thermal stability.
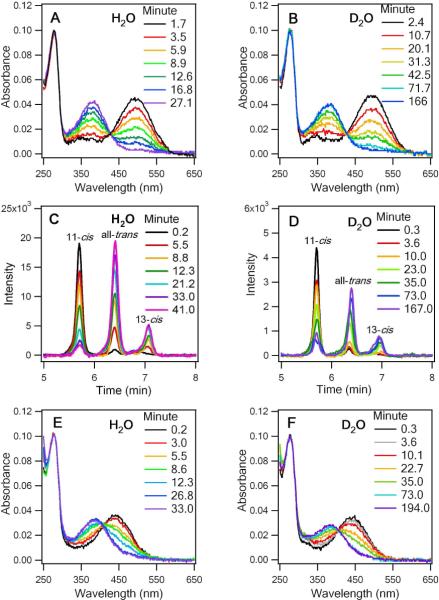
To examine the kinetics of thermal decay, we measured differences in the UV-vis absorption of rhodopsin at appropriate time intervals at 59°C to determine the rates of thermal decay in H2O and D2O (Figure 1A and B). Absorption at 280 nm corresponds to the aromatic amino acids of opsin, while the signal at 500 nm corresponds to rhodopsin with the 11-cis retinal chromophore covalently bound to opsin by a PSB. At t > 0, the intensity of the 500 nm peak decreases, while a peak appears at 380 nm with increasing intensity. This 380 nm peak could be attributed to free 11-cis retinal, free all-trans retinal, and/or all-trans retinal bound to opsin. Because only dark state rhodopsin contributes to OD500, a decrease of OD500 reflects the thermal decay of rhodopsin and is fitted to a single-exponential function to obtain a half-life of 7 ± 1 min in H2O and 21 ± 3 min in D2O as indicated in Figure 2A.
Figure 1.
UV-Vis spectra of the thermal decay of WT rhodopsin (WT rho) at different time points at 59°C in (A) H2O and (B) D2O. HPLC traces of retinal extractions at different time points: (C) H2O and (D) D2O. UV-Vis spectra of the acid-denatured WT rhodopsin: (E) H2O and (F) D2O.
Figure 2.
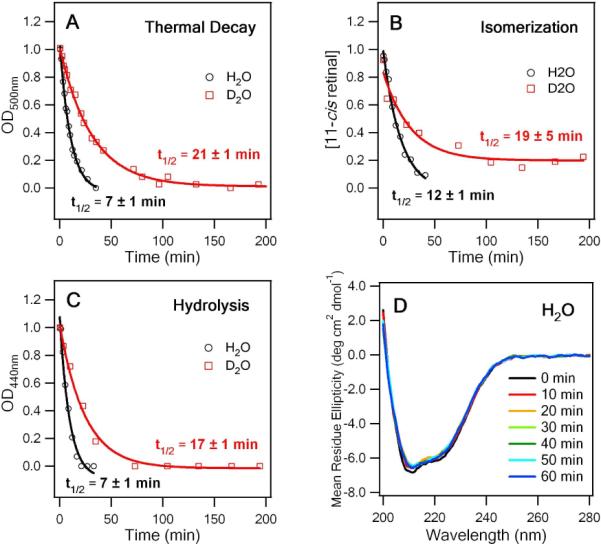
(A) Rates of thermal decay for WT rho D2O and H2O determined by OD500 at 59°C. (B) Rates of thermal isomerization for WT rho in D2O and H2O at 59°C determined by the fraction of 11-cis retinal. (C) The rate of hydrolysis for WT rho determined by the decay of OD440. (D) The CD spectra of WT rho incubated at 59°C for different time intervals.
Subsequently, to quantify the rates of thermal isomerization, retinal was extracted from samples of rhodopsin incubated at 59°C at different time intervals and analyzed by HPLC. The 11-cis peak decreases whereas the all-trans peak increases, confirming that 11-cis retinal isomerizes to all-trans retinal during thermal decay (Figures 1C & D). The chromatographs also indicate the appearance of a minor 13-cis retinaloxime that could be due to spontaneous thermal isomerization from all-trans and/or 11-cis retinal during incubation at 59°C. We calculated the fraction of 11-cis retinal in each sample (see Supporting Information) and determined that thermal isomerization of 11-cis retinal in rhodopsin occurs with a half-life of 19 ± 5 min in D2O, which is approximately two-fold faster than that in H2O (12 ± 1 min, Figure 2B). Incidentally, an experimental control which quantified the isomerization of 11-cis retinal in the absence of protein yielded an average half-life of 613 ± 170 min, forty times slower than that of retinal tethered to opsin. Thus, we conclude that the observed thermal isomerization occurs within the retinal binding site and not after dissociation from the opsin protein.
To study the kinetics of PSB hydrolysis, we applied an acid-denaturation assay, in which the pH of each sample was lowered to 1–2 to denature the opsin protein.13 At the earliest time point (t = 1.0 min), the spectrum predominately peaks at 440 nm, corresponding to retinal that remained covalently attached to opsin via a PSB (Figure 1E). Upon thermal decay of rhodopsin, OD440 decreases, while OD380 increases, indicating that the PSB was hydrolyzed such that retinal was no longer linked to the protein to give a 380-nm UV absorption. To quantify the extent of PSB hydrolysis, we plotted each absorption spectrum as a function of frequency and fit it as a sum of two Gaussian functions with peaks at 380 nm and 440 nm to obtain half lives of 7 ± 1 min and 17 ± 1 min for H2O and D2O, respectively (Figure 2C). This result provides evidence to support that hydrogen bonds play a role in the hydrolysis process.
A recent study has proposed that thermal denaturation of rhodopsin forces the retinal to isomerize to the all-trans configuration and to leave the binding pocket.9 To verify whether the thermal isomerization is associated with denaturation of the protein, we used circular dichroism (CD) spectroscopy to investigate the extent of unfolding of the rhodopsin secondary structure during thermal decay. The spectra (Figure 2D) reveal characteristic contributions from both α-helices located primarily in the transmembrane region of rhodopsin and β-sheets on the extracellular side.14 Beyond the half life of the thermal decay process (> 30 min), the mean residue ellipticity at 210 nm changes less than 5%, roughly equal to the signal fluctuation in the spectral region, suggesting that changes in secondary structure neither contribute significantly to the thermal decay process nor trigger the thermal isomerization of 11-cis to all-trans retinal. However, it remains possible that the denaturation of rhodopsin could occur at a level beyond secondary structure, such as helical translocations in tertiary structure, to result in compromised thermal stability and function, a notion which aligns with the proposal offered by del Valle et al.9
In this study, we report that the thermal decay process of rhodopsin is not attributed to a single thermal event, as previously suggested, but instead involves the simultaneous hydrolysis of PSB and thermal isomerization of 11-cis to all-trans retinal. In addition, investigations of the unfolding of rhodopsin by CD and of the rate of thermal isomerization of 11-cis retinal free in solution have shown that the observed thermal isomerization occurs in the binding site of rhodopsin with secondary structure intact. Finally, we have demonstrated that the rates of thermal decay, isomerization, and hydrolysis were significantly slower in D2O, suggesting that hydrogen bonds, which are stronger for deuterium,15 are involved in the rate-determining step in the thermal decay process and thereby influence the thermal stability of rhodopsin.
On the basis of structural analysis, Schertler and coworkers proposed that an hydrogen-bonding network is important for stabilizing rhodopsin.16 We now extend the proposal by speculating that this network influences the kinetics of thermal processes, including thermal isomerization and hydrolysis in rhodopsin, and may be implicated in certain visual ailments such as retinitis pigmentosa and congenital night blindness. In order to explore the underlying mechanisms behind the progression of these diseases, it is important to examine the role of the hydrogen-bonding network on rhodopsin tertiary structure and on the kinetics of thermal decay of rhodopsin. Studies of rhodopsin with mutations in the hydrogen-bonding network are currently underway to achieve this goal.
Supplementary Material
Acknowledgement
We thank Prof. Thomas P. Sakmar and Dr. Shixin Ye (The Rockefeller University) for the support and contribution in the pilot study of this research.
Footnotes
Supporting Information Available: Experimental details (PDF) are available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Menon ST, Han M, Sakmar TP. Physiol. rev. 2001;81:1659–1688. doi: 10.1152/physrev.2001.81.4.1659. [DOI] [PubMed] [Google Scholar]
- (2).Barlow RB, Birge RR, Kaplan E, Tallent JR. Nature. 1993;366:64–66. doi: 10.1038/366064a0. [DOI] [PubMed] [Google Scholar]
- (3).Aho AC, Donner K, Hyden C, Larsen LO, Reuter T. Nature. 1988;334:348–350. doi: 10.1038/334348a0. [DOI] [PubMed] [Google Scholar]
- (4).Birge RR, Barlow RB. Biophys. Chem. 1995;55:115–126. doi: 10.1016/0301-4622(94)00145-a. [DOI] [PubMed] [Google Scholar]
- (5).Baylor D, Matthews G, Yau K-W. J. Physiol. 1980;309:591–621. doi: 10.1113/jphysiol.1980.sp013529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Baylor DA, Nunn BJ, Schnapf JL. J. Physiol. 1984;357:575–607. doi: 10.1113/jphysiol.1984.sp015518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Janz JM, Farrens DL. J. Biol. Chem. 2004;279:55886–55894. doi: 10.1074/jbc.M408766200. [DOI] [PubMed] [Google Scholar]
- (8).Janz JM, Fay JF, Farrens DL. J. Biol. Chem. 2003;278:16982–16991. doi: 10.1074/jbc.M210567200. [DOI] [PubMed] [Google Scholar]
- (9).Del Valle LJ, Ramon E, Bosch L, Manyosa J, Garriga P. Cell. Mol. Life Sci. 2003;60:2532–2537. doi: 10.1007/s00018-003-3113-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau EM, Shichida Y. Proc. Natl. Acad. Sci. USA. 2002;99:5982–5987. doi: 10.1073/pnas.082666399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Yan ECY, Kazmi MA, Ganim Z, Hou JM, Pan D, Chang BSW, Sakmar TP, Mathies RA. Proc. Natl. Acad. Sci. USA. 2003;100:9262–9267. doi: 10.1073/pnas.1531970100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Yan ECY, Epps J, Lewis JW, Szundi I, Bhagat A, Sakmar TP, Kliger DS. J. Phys. Chem. C. 2007;111:8843–8848. [Google Scholar]
- (13).Tsutsui K, Imai H, Shichida Y. Biochemistry. 2007;46:6437–6445. doi: 10.1021/bi7003763. [DOI] [PubMed] [Google Scholar]
- (14).Crescitelli F, Mommaerts WFHM, Shaw TI. Proc. Natl. Acad. Sci. USA. 1966;56:1729–1734. doi: 10.1073/pnas.56.6.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Legon AC, Millen DJ. Chem. Phys. Lett. 1988;147:484–489. [Google Scholar]
- (16).Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. J. Mol. Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.