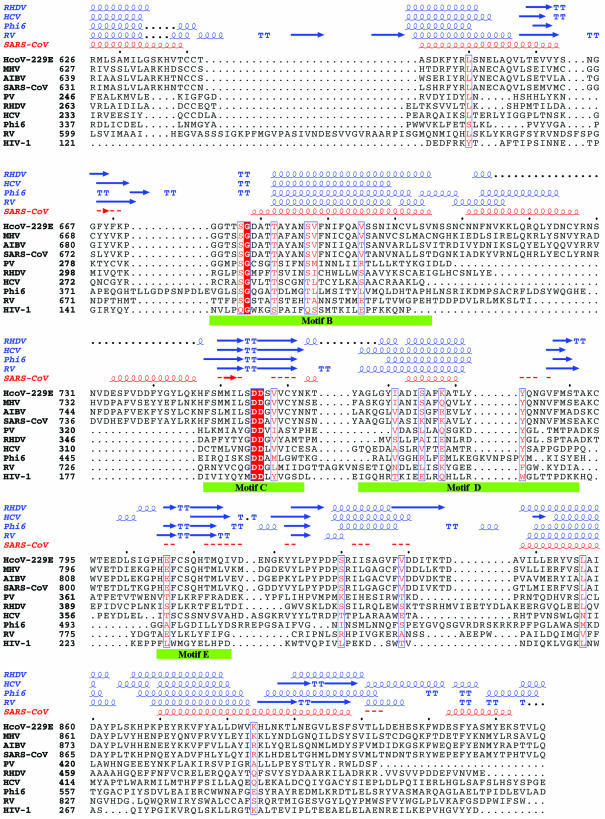
Figure 1.
Sequence alignment of the RdRp of SARS-CoV with those of representatives of the other three classes of coronaviruses and of five RNA viruses with known crystal structures. The representative coronaviruses are: group I, human coronavirus 229E (HcoV-229E; NCBI accession no. NC_002645); group II, murine hepatitis virus (MHV; NCBI accession no. NC_001846); and group III, avian infectious bronchitis virus (AIBV; NCBI accession no. NC_001451). The five RNA viruses are poliovirus 1 strain Mahoney (PV; PDB code 1RDR), rabbit hemorrhagic disease virus (RHDV; PDB code 1KHV), hepatitis C virus (HCV; PDB code 1QUV), reovirus (RV; PDB codes 1N35 and 1N1H) and bacteriophage φ6 (Phi6; PDB codes 1HI0 and 1HI1). HIV-1 RT (HIV-1; PDB code 1RTD), a widely studied RNA-dependent and DNA-dependent polymerase, is also included in the comparison. The sequence in the palm subdomain and regions containing the conserved motifs (highlighted with green bars) can be aligned confidently among different viral RdRps and HIV-1 RT. However, the sequence in the fingers and thumb subdomains is less conserved between SARS-CoV RdRp and other viral RdRps, and the structure in those subdomains also varies substantially among the known RdRp structures. Thus, the sequence alignment and the structural model in these regions are less reliable. Invariant residues are highlighted in a shaded red box, and conserved residues are in red. The secondary structures of RHDV, HCV, RV and φ6 polymerases extracted from the corresponding structures and the predicted secondary structure of SARS-CoV RdRp are shown above the sequence alignment. α-Helices are shown as spirals and β-strands as arrows. The alignment was drawn with ESPript (66).