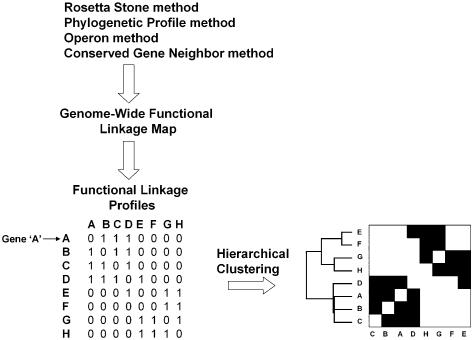
Figure 3.
Schematic of the methods used in this study. The Rosetta Stone, Phylogenetic Profile, Operon and Conserved Gene Neighbor methods were employed to identify functionally linked proteins throughout the M.tuberculosis genome. Functional linkages inferred by two or more computational methods were used to construct a genome-wide functional linkage map. The map was then converted into separate bit vectors representing the functional linkage profile of each gene in the map. Hierarchical clustering resulted in a reordering of the genes, clustering genes with similar functional linkage profiles.