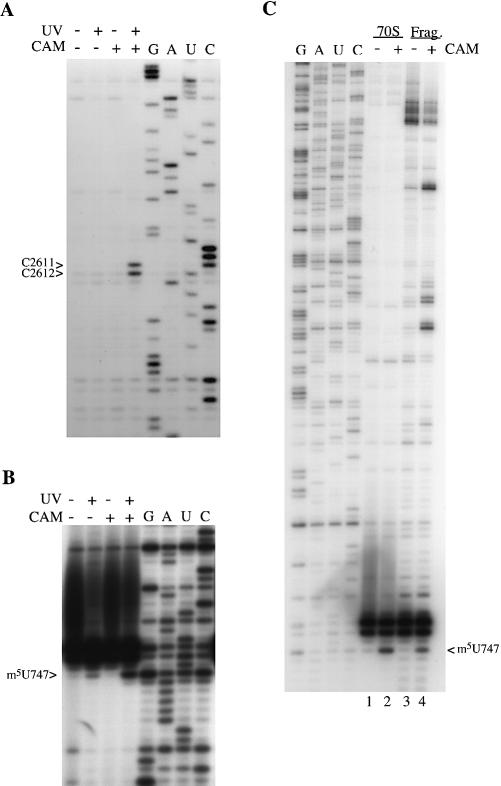
Figure 2.
Chloramphenicol-dependent, UV-induced modifications in E.coli 23S rRNA. (A and B) Ribosomes (0.15 µM) were incubated with (+) and without (–) chloramphenicol (1.2 mM) for 20 min at 37°C, and with (+) and without (–) UV irradiation at 365 nm for 30 min. Ribosomal RNA was isolated and analyzed by primer extension with primers Ec2654 (A) and Ec770 (B). (C) RNase H analysis of the modification at 747. Ribosomes (0.5 µM) were incubated with (+) and without (–) chloramphenicol (1.2 mM) for 20 min at 37°C, irradiated at 365 nm for 30 min, followed by isolation of rRNA. A small portion of each sample (70S) was analyzed directly by extension from primer Ec770. The remainder of each sample was treated with RNase H in the presence of oligonucleotides Ec617 and Ec820. The released fragment (Frag.) was gel-purified and subjected to primer extension using primer Ec770. In (B) and (C), 23S rRNA from the E.coli IB10 strain lacking the m1G745 methyltransferase and the strong stop at G745 was used for the sequencing tracks (43). The rRNA modification-induced stop is displaced by one base below the corresponding stop in the sequencing tracks (G, A, U and C).