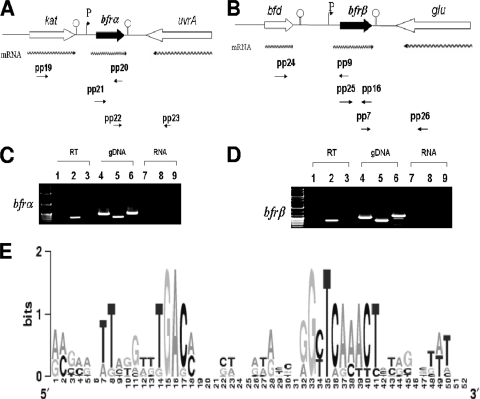
FIG. 1.
Transcriptional organization analysis and transcription start site determination for bfrα and bfrβ in P. putida KT2440. (A) Organization of the bfrα neighborhood in P. putida KT2440. Wavy arrows indicate predicted transcripts, and straight arrows show primer targets used for RT-PCR. (B) Organization of the bfrβ neighborhood in P. putida KT2440. Wavy arrows indicate predicted transcripts, and straight arrows show primer targets used for RT-PCR. (C) PCR detection of the upstream region of bfrα (spanning the kat-bfrα) by using primer pair pp19/pp20 (see Table 2 for details on all primers) (lanes 1, 4, and 7), bfrα by using primer pair pp21/pp20 (lanes 2, 5, and 8), and the bfrα-uvrA junction by using primer pair pp22/pp23 (lanes 3, 6, and 9). (D) PCR detection of the upstream region of bfrβ (spanning the bfd-bfrβ junction) by using primer pair pp24/pp9 (lanes 1, 4, and 7), bfrβ by using primer pair pp25/pp16 (lanes 2, 5, and 8), and the bfrβ-glu junction by using primer pair pp7/pp26 (lanes 3, 6, and 9). For C and D, PCR templates were genomic DNA (gDNA) (positive control), cDNA (RT), or non-reverse-transcribed RNA (RNA) (negative control). (E) Logo of putative promoter motifs for the bfrα and bfrβ genes in selected fluorescent pseudomonads. The nonencoding regions within 100 bp upstream of the bfrα and bfrβ translation start codons were first manually scanned, aligned with ClustalW, and preceded with WebLogo (13). The selected bfrα and bfrβ genes are from P. putida KT2440 (GenBank accession number AE015451), P. aeruginosa PAO1 (accession number AE004091), Pseudomonas fluorescens PfO-1 (accession number CP000094), and Pseudomonas syringae pv. syringae B728a (accession number CP000075) (see Fig. S2 in the supplemental material).