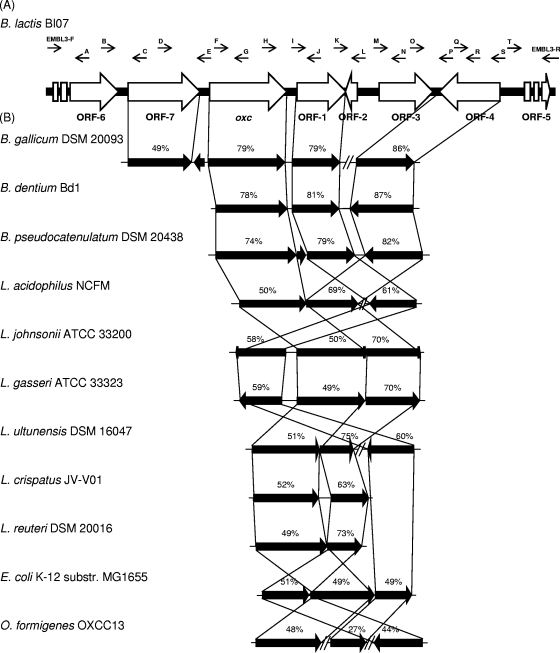
FIG. 2.
Genetic organization around the oxc gene in B. animalis subsp. lactis BI07 (A) and comparison with the corresponding loci in bifidobacteria, lactobacilli, E. coli, and O. formigenes (B). A through T, primers A through T used for primer walking (Table 1). Each arrow indicates an ORF, and the length of the arrow is proportional to the length of the predicted ORF. Levels of amino acid identity, expressed as percentages, are shown. The organisms used were B. gallicum DSM 20093 (accession no. NZ_ABXB00000000), B. dentium Bd1 (54), B. pseudocatenulatum DSM 20438 (accession no. NZ_ABXX00000000), L. acidophilus NCFM (3), Lactobacillus johnsonii ATCC 33200 (accession no. NZ_ACGR00000000), Lactobacillus gasseri ATCC 33323 (36), Lactobacillus ultunensis DSM 16047 (accession no. NZ_ACGU00000000), Lactobacillus crispatus JV-V01 (accession no. NZ_ACKR00000000), Lactobacillus reuteri DSM 20016 (accession no. NC_009513), E. coli K-12 MG1655 (accession no. NC_000913), and O. formigenes OXCC13 (accession no. NZ_ACDQ00000000).