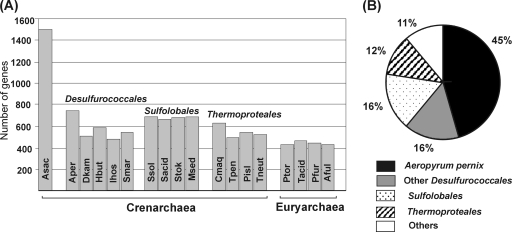
FIG. 1.
Comparisons of proteomes of A. saccharovorans and other archaea. (A) The numbers of A. saccharovorans protein-encoding genes present in the genomes of other archaea. Genes were considered present in a pair of genomes if the region of similarity covered >70% of the corresponding protein with an E value of <10−10. Abbreviations: Asac, A. saccharovorans; Aper, Aeropyrum pernix; Dkam, Desulfurococcus kamchatkensis; Hbut, Hyperthermus butylicus; Ihos, Ignicoccus hospitalis; Smar, Staphylothermus marinus; Ssol, Sulfolobus solfataricus; Sacid, Sulfolobus acidocaldarius; Stok, Sulfolobus tokodaii; Msed, Metallosphaera sedula; Cmaq, Caldivirga maquilingensis; Tpen, Thermofilum pendens; Pisl, Pyrobaculum islandicum; Tneut, Thermoproteus neutrophilus; Ptor, Picrophilus torridus; Tacid, Thermoplasma acidophilum; Pfur, Pyrococcus furiosus; Aful, Archaeoglobus fulgidus. (B) The best BLASTP hits of A. saccharovorans proteins. Significant BLASTP hits were detected for 1,218 of 1,499 proteins found in the A. saccharovorans genome. Proteins unique to A. saccharovorans were excluded from this analysis.