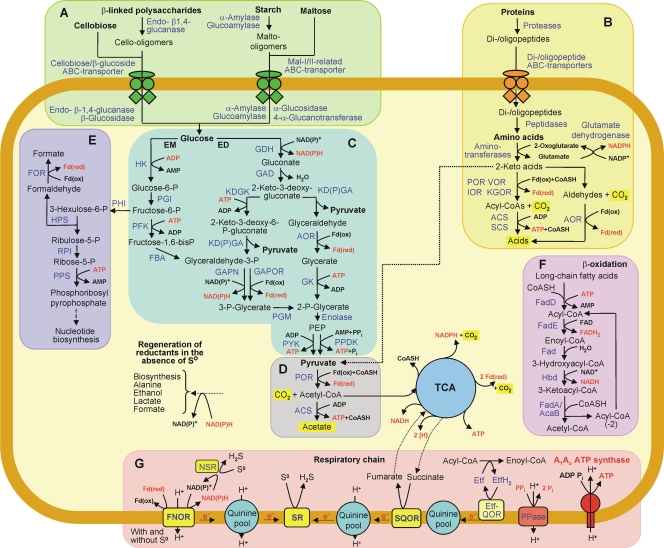
FIG. 2.
Overview of catabolic pathways encoded by the A. saccharovorans genome. Substrates utilized are in boldface, enzymes and proteins encoded by genes identified on the genome are in blue, and energy-rich intermediate compounds are in red. Panels: A, utilization of carbohydrates; B, utilization of proteins; C, glycolysis (EM and ED pathways); D, pyruvate degradation; E, pentose phosphate synthesis; F, β-oxidation of long-chain fatty acids; G, formation of proton motive force coupled with ATP generation. TCA, oxidative tricarboxylic acid cycle. Abbreviations: POR, pyruvate:ferredoxin oxidoreductase; VOR, 2-ketoisovalerate:ferredoxin oxidoreductase; IOR, indolepyruvate:ferredoxin oxidoreductase; KGOR, 2-ketoglutarate:ferredoxin oxidoreductase; ACS, acetyl-CoA synthetase; SCS, succinyl-CoA synthetase; AOR, aldehyde:ferredoxin oxidoreductases; HK, ADP-dependent hexokinase; PGI, phosphoglucose isomerase; PFK, ADP-dependent phosphofructokinase; FBA, fructose-1,6-bisphosphate aldolase; GAPOR, glyceraldehyde-3-phosphate:ferredoxin oxidoreductase; GAPN, nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase; PGM, phosphoglycerate mutase; PYK, pyruvate kinase; PPDK, pyruvate phosphate dikinase; PEP, phosphoenolpyruvate; GDH, glucose dehydrogenase; GAD, gluconate dehydratase; KDGK, 2-keto-3-deoxy-gluconate kinase; KD(P)GA, 2-keto-3-deoxy-(6-phospho)gluconate aldolase; GK, glycerate kinase; PHI, 6-phospho-3-hexuloisomerase; HPS, 3-hexulose-6-phosphate synthase; RPI, ribose-5-phosphate isomerase; PPS, phosphoribosyl pyrophosphate synthase; FOR, formaldehyde:ferredoxin oxidoreductase; Fd(ox), oxidized ferredoxin; Fd(red), reduced ferredoxin; FNOR, ferredoxin:NAD(P)+ oxidoreductase complex; NSR, NAD(P)H:elemental sulfur oxidoreductase; PPase, H+-translocating pyrophosphatase; SQOR, succinate:quinone oxidoreductase complex; SR, sulfur reductase complex; Etf, electron transfer flavoprotein; Etf-QOR, electron transfer protein-quinone oxidoreductase; CoASH, coenzyme A; FAD, flavin adenine dinucleotide; FADH2, reduced FAD.