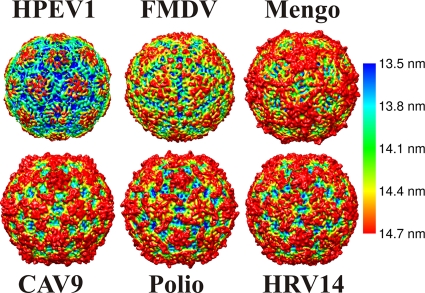
FIG. 3.
Comparison of the icosahedrally symmetric 0.85-nm-resolution HPEV1 reconstruction with the atomic models of other picornaviruses. HPEV1, FMDV (Protein Data Bank [PDB] code 1bbt), mengovirus (PDB 2mev), CAV9 (PDB 1d4m), poliovirus (1hxs), and HRV14 (4rhv). The X-ray models have been filtered to 0.85-nm resolution, and all models are rendered as isosurface representations at 2 standard deviations above the mean, viewed down a 2-fold axis of symmetry. This comparison demonstrates the diversity of picornavirus surface features as well as the unique features of HPEV1. The models have been colored using radial-depth cueing in CHIMERA (bar, 13.5- to 14.7-nm radius) (60).