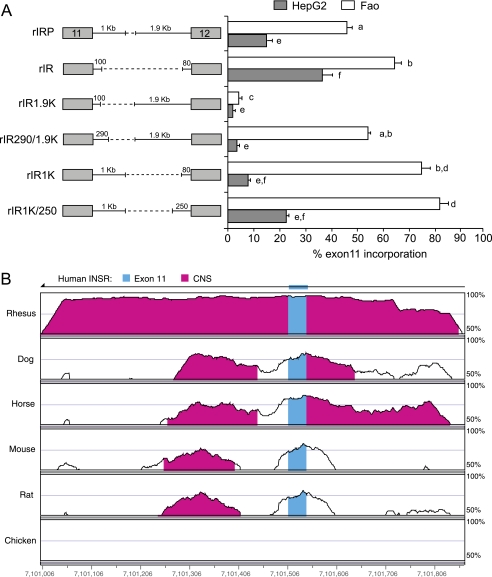
FIGURE 3.
Comparison of the rat and human INSR genes identifies a conserved downstream splicing element. A, schematic of rat IR minigene deletion mutants transfected into HepG2 and Fao cells. Numbers above the lines indicate length of rat intron sequence. Percent of exon 11 inclusion in HepG2 (gray bar) and Fao cells (white bar) is shown in the bar graph on the right. Results are derived from three independent experiments and are given as mean ± S.E. Statistical significance was analyzed by ANOVA for each cell line independently. Bars with the same letter are not significantly different (p < 0.05). B, vista alignment plot of human, rhesus monkey, dog, horse, mouse, rat, and chicken genomic sequences covering 800 nucleotides, including exon 11 and the ends of introns 10 and 11. Exonic structure of the human INSR gene is shown at the top. The gene reads from right to left as indicated by the arrow at the top, so intron 11 is to the left of exon 11. Blue areas indicate exon 11, and magenta areas indicate conserved nucleotide sequence (CNS) between the seven genomes. Conservation in these regions exceeds 50%. The chicken INSR gene does not contain an exon 11 and only encodes a single insulin receptor.