Abstract
Methyl-coenzyme M reductase (MCR) from methanogenic archaea catalyzes the final step of methane formation, in which methyl-coenzyme M (2-methylthioethane sulfonate, methyl-SCoM) is reduced with coenzyme B (N-7-mercaptoheptanolyl-threonine phosphate, CoBSH) to form methane and the heterodisulfide CoBS-SCoM. The active dimeric form of MCR contains two Ni(I)-F430 prosthetic groups, one in each monomer. This manuscript describes studies of the reaction of the active Ni(I) state of MCR (MCRred1) with BES (2-bromoethanesulfonate) and CoBSH or its analog, CoB6SH (N-6-mercaptohexanolyl-threonine phosphate), by transient kinetic measurements using EPR and UV-visible spectroscopy and by global fits of the data. This reaction is shown to lead to the formation of three intermediates, the first of which is assigned as an alkyl-Ni(III) species that forms as the active Ni(I)-MCRred1 state of the enzyme decays. Subsequently, a radical (MCRBES radical) is formed that was characterized by multifrequency electron paramagnetic resonance (EPR) studies at X (~ 9 GHz)-, Q (~35 GHz)- and D (~135 GHz)-bands and by electron-nuclear double resonance (ENDOR) spectroscopy. The MCRBES radical is characterized by g-values at 2.00340 and 1.99832 and includes a strongly coupled non-exchangeable proton with a hyperfine coupling constant of 50 MHz. Based on transient kinetic measurements, the formation and decay of the radical coincides with a species that exhibits absorption peaks at 426 nm and 575 nm. Isotopic substitution, multifrequency EPR and ENDOR spectroscopic experiments rule out the possibility that MCRBES is a tyrosyl radical and indicate that if a tyrosyl radical is formed during the reaction, it does not accumulate to detectable levels. The results provide support for a hybrid mechanism of methanogenesis by MCR that includes both alkyl-Ni and radical intermediates.
Keywords: Methane, methyl-Coenzyme M, methanogenesis, nickel, coenzyme F430, EPR spectroscopy, ENDOR spectroscopy, enzyme kinetics, radical
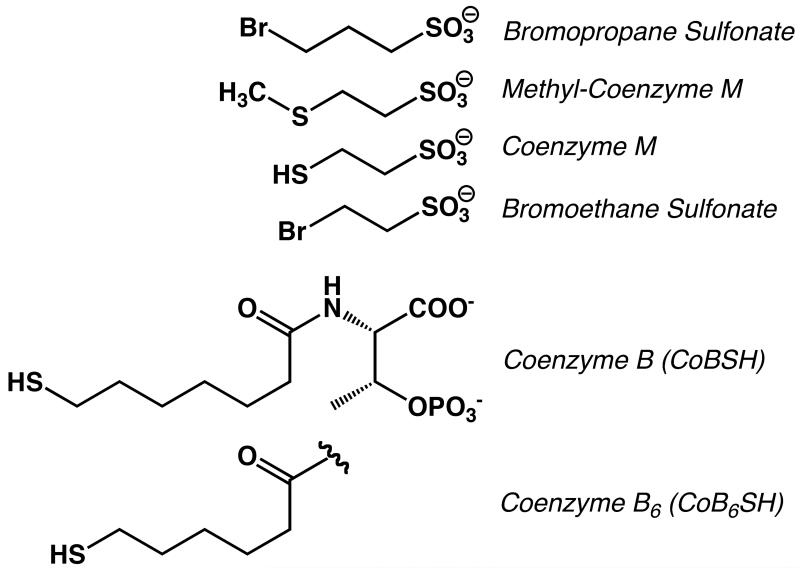
Methyl-coenzyme M reductase (MCR) from methanogenic archaea catalyzes the final step of methane formation, in which methyl-coenzyme M (methyl-SCoM, 2-methylthioethane sulfonate) is reduced with coenzyme B (CoBSH, N-7-mercaptoheptanolyl-threonine phosphate) to form methane and the heterodisulfide CoBS-SCoM according to eq1 (1, 2). The structures of these substrates and analogs described in this paper are shown in Scheme 1.
| (1) |
Scheme 1.
Structures of HSCoM analogs.
X-ray crystallographic studies have revealed that MCR is composed of three different subunits in an (αβγ)2 structure with an F430 molecule in each of the two active sites (3). Coenzyme F430, a redox-active nickel tetrahydrocorphin cofactor, noncovalently binds to MCR at the bottom of a 30 Å long hydrophobic well that accommodates the two substrates and shields the reaction from solvent. The phosphate group of CoBSH is positioned by ionic interactions with MCR residues located halfway down this channel with its thiol group located 8.7 Å from the nickel. Methyl-SCoM binds in the pocket close to F430 such that the thioether sulfur is positioned above the nickel and about 6.4 Å from the sulfur of CoBSH. There exist various oxidation and ligation states of F430 in MCR, including EPR-active forms (MCRred1, MCRred2, MCRox1 and MCRox2) (4), and EPR-silent Ni(II) forms (MCRsilent, MCRred1-silent and MCRox1-silent) (3, 5, 6). MCRred1 with Ni(I)-F430 is known to be the active form of MCR and exhibits characteristic EPR spectra with the following g-values: g⊥ = 2.065, g∥= 2.24 (7-9).
The active MCRred1 state has been shown to undergo a variety of reactions, some of which lead to EPR-detectable species. In the presence of HSCoM and CoBSH, the MCRred1 state is partly converted into the MCRred2 state, which has a rhombic Ni(I)-type EPR signal (10). The reaction of MCRred1 with various alkyl bromides leads to the formation of organometallic alkyl-Ni species. For example, the reaction of MCRred1 with 3-bromopropanesulfonate (BPS), the most potent known inhibitor of methanogenesis (11), generates MCRPS, (originally termed MCRBPS (12)) which is an organometallic Ni(III)/propyl sulfonate species in resonance with a Ni(II)/propyl sulfonyl radical (13). The EPR spectrum of MCRPS exhibits g-values at 2.108, 2.112, and 2.219, with most (~75%) of the spin density located in the nickel 3d(x2-y2) orbital (13). Similarly, when MCRred1 is reacted with methyl iodide or methyl bromide, an organometallic methyl-Ni(III) species is formed (MCRMe) (14, 15). Similar organo-Ni(III) species are generated when MCRred1 is reacted with various brominated acids (16). Both MCRMe and MCRPS can be converted to the active MCRred1 state under certain conditions; for example by reaction with thiolates like HSCoM (12, 16, 17).
While MCRred1 reacts with BPS to generate an organo-Ni(III) species, the reaction of MCRred1 with 2-bromoethanesulfonate (BES) in the presence of CoBSH generates a radical (MCRBES radical), characterized by a doublet EPR signal (18). BES is a reversible inhibitor that is competitive with respect to methyl-SCoM (19, 20) and has also been used to inhibit methanogenesis within the bovine rumen (21). Several other methyl-SCoM analogs, (3-bromopropionate, cyano-coenzyme M, seleno-coenzyme M and trifluoromethyl-coenzyme M), also have been shown to induce the doublet radical EPR signal when they are reacted with MCRred1 in the presence of CoBSH (18). The studies described in the current manuscript were focused on obtaining a better understanding of the nature of the reaction of MCR with BES. Prior studies indicated that reactive haloalkyl compounds that are the size of HSCoM generate radicals and those that are the size of methyl-SCoM (and larger) generate organometallic species, suggesting that subtle changes in length of the reactive inhibitor (e.g., BPS versus BES) reflect the extreme selectivity in the MCR active site. However, as shown here, the reaction with BES also generates an organometallic intermediate that rapidly converts to the characteristic MCRBES radical. Therefore, when comparing various haloalkanes, the nature of the intermediates actually may not change; instead the rates of formation and decay of the organometallic, radical, and other intermediates vary, thus altering the amount of these species that can accumulate.
Three types of mechanisms for MCR-based catalysis have been proposed, Mechanism I involving an organometallic methyl-Ni intermediate (6, 22), Mechanism II involving a methyl radical (23) and Mechanism III, which involves methyl-Ni and interactions between the substrate and the tetrapyrrole ring of F430 (24). Mechanism I, proposed based on the crystal structure (3) and mechanistic work with F430 model complexes (25-27), involves nucleophilic attack of Ni(I)-MCRred1 on the methyl group of methyl-SCoM to generate a methyl-Ni intermediate (17). Mechanism III starts with protonation of coenzyme F430 (42), which promotes reductive cleavage of the methyl-SCoM thioether bond generating a nickel center that is coordinated by −CH3 and −SCoM anions and two of the tetrapyrrole nitrogens. The CoBS− anion is then proposed to react with −SCoM to form the heterodisulfide product and methyl-Ni(I). Formation of methane requires the proton that was initially donated to coenzyme F430. Mechanism I is supported by the characterization of various alkyl-Ni species that have been generated by the reaction of MCRred1 with 3-bromopropionate (BPS) and other alkyl halides (14, 16, 28). In addition, MCRPS and other alkyl-Ni(III) species can be converted to the active MCRred1 state by reacting with organic thiolates, which forms a thioether product (17). Mechanism II is based on density functional theory (DFT) computations and proposes that Ni(I) attacks the sulfur atom of methyl-SCoM, promoting homolytic cleavage of the methyl-sulfur bond and generating a MCRox1-like Ni(III)-SCoM complex and a methyl radical, which abstracts a hydrogen atom from CoBSH to generate methane (23, 29, 30).
Mechanisms I-III remain viable because neither the methyl-Ni(III) intermediate nor the Ni(III)-SCoM species has been observed upon reaction of MCRred1 with the native substrate methyl-SCoM. In fact, no spectroscopic changes have been reported when MCRred1 is reacted with the native substrates either separately or together, regardless of the order of addition or the concentration of the substrates. Thus, in order to probe the reaction mechanism, substrate analogs have been used. Among these, 2-bromoethanesulfonate (BES) is similar in size to methyl-SCoM and contains an excellent leaving group (Br−), promoting a rapid reaction with MCR.
Here, rapid kinetic and spectroscopic experiments were performed to better understand the reaction of MCRred1 with BES in the presence of CoBSH or an analog, N-6-mercaptohexanolyl-threonine phosphate (CoB6SH). The combined results indicate that formation of the MCRBES radical is a three-step reaction, and that the first step involves formation of a transient alkyl-Ni(III) species that rapidly decays to form the doublet radical species. The UV-visible spectra indicate that the radical has long wavelength absorption (~550 nm). Isotopic substitution coupled with EPR and ENDOR experiments also rule out the possibility that the radical originates from one of the two Tyr residues located directly above the active site Ni center.
MATERIALS AND METHODS
Materials
Methanothermobacter marburgensis was obtained from the Oregon Collection of Methanogens (Portland, OR) catalog as OCM82. CoBSH and CoB6SH were synthesized from 7-bromoheptanoic acid and 6-bromohexanoic acid, respectively as described previously (28, 31, 32). The purities of CoBSH and CoB6SH were ascertained to be >98% by 1H NMR. [2H6]-p-hydroxyphenylacetic acid was prepared from p-hydroxyphenylacetic acid as described (33). The yield was measured by mass spectrometry with the following distribution of deuterium: 80% of 2H6-, 18% of 2H5- and 2% of 2H4- p-hydroxyphenylacetic acid. Other reagents were purchased from Sigma-Aldrich. N2 (99.98%), H2/CO2 (80%/20%), and ultra high purity H2 (99.999%) were obtained from Cryogenic Gases (Detroit, MI).
Protein purification
The purification of MCRred1 from Mt. marburgensis was performed under strictly anaerobic conditions, as described previously (17). According to the UV-visible spectrum (representative spectra are shown below), 50 – 70% of MCRred1 was typically generated. MCRred1, which was universally labeled with 2H6-Tyr, was generated by culturing Mt. marburgensis in the presence of 3 mM [2H6]-p-hydroxyphenylacetic acid, 0.1 mM phenylacetate and 0.02 mM indoleacetate, which are precursors of Tyr (M. marburgensis does not appear to import Tyr) (33). Peptide hydrolysis followed by mass spectrometric and amino acid analyses were used to determine the amount of [2H6]-p-hydroxyphenylacetic acid incorporated into the tyrosine residues in MCR. For amino acid analysis, MCR was hydrolyzed using 6 M HCl for 24 h at 110 °C, and then the amino residues were treated with bis-trimethylsilyl-trifluoroacetamide (BSTFA) and acetonitrile (1:1, v/v) for 1 h at 100 °C. These trimethylsilyl derivatives were analyzed using a Finnigan Trace GC/MS. O-acetylation of tyrosine in MCR was obtained through treatment of 130 μM MCR with 280 mM N-acetylimidazole on ice for 2.5 h. The excess N-acetylimidazole was removed with a molecular weight cut-off centrifugal filter.
Spectroscopy of MCR
UV-visible spectra of MCR were recorded in the anaerobic chamber using a diode array spectrophotometer (model DT 1000A, Analytical Instrument Systems, Inc., Flemington, NJ). X-band EPR spectra were recorded on a Bruker EMX spectrometer (Bruker Biospin Corp., Billerica, MA), equipped with an Oxford ITC4 temperature controller, a Hewlett-Packard model 5340 automatic frequency counter, and Bruker gaussmeter. Spin concentration was determined by double integration of the sample spectrum obtained under non-saturating conditions and comparing to that of 1 mM copper perchlorate standard. All samples for EPR spectroscopy were prepared in 50 mM Tris buffer, pH 7.6, in a Vacuum Atmospheres anaerobic chamber (Hawthorne, CA) maintained under nitrogen gas at <1 ppm of oxygen.
CW 35 GHz (“Q-band”) EPR and ENDOR (34) spectra were recorded on a modified Varian E-110 spectrometer equipped with a helium immersion dewar at 2 K under “rapid passage” (35) conditions using 100 kHz field modulation (36). Under these conditions, the EPR spectrum appears as an absorption lineshape, rather than in first derivative mode. Pulsed 35 GHz echo-detected EPR and ENDOR spectra were obtained at 2 K on a locally constructed spectrometer (37). For the echo-detected EPR spectra, the signal intensity of a standard Hahn two-pulse spin-echo is monitored as a function of field sweep. Multiple echo intensities are averaged and the EPR spectrum can be recorded at various repetition rates to probe the relaxation behavior of multiple paramagnetic sites within a sample. The lineshape of such an echo-detected EPR spectrum may show distortions from that expected for an ideal powder pattern as the phase of the spin echo can vary during the data acquisition, due to instrumental limitations. More importantly, the T1 is not a constant over the EPR envelope, even for a single paramagnetic site. As it is also not experimentally feasible to wait enough multiples of the longest T1 between pulse cycles to allow full recovery of the magnetization (ideally, a repetition rate > 10 T1), the degree of saturation and hence the intensity varies across the spectrum, distorting it. Echo-detected EPR spectra necessarily appear as the absorption lineshape, so that the 35 GHz CW and pulsed EPR spectra have the same general appearance.
For the CW ENDOR spectra (34), two methods were employed: sweeping the rf (in either direction), as is commonly done (36), or by random hopping of the rf using a sample time, delay time, and rf time, as has been recently developed (38). This latter method avoids many of the lineshape artifacts found in swept CW ENDOR spectra (39), but generally gives a lower signal/noise ratio. For the pulsed 35 GHz ENDOR spectra, the Mims three-pulse ENDOR sequence, tmw-τ-tmw-T(rf)-tmw-τ-echo (34, 40) was employed, where tmw is the π/2 microwave pulse length (typically 50 ns), τ is a preparation (or delay) time (typically 500 ns), and T(rf) is the time during which the rf is applied (typically 60 μs). The Mims sequence has the property that its ENDOR intensities follow the relationship, I(A) ~ 1 - cos(2πAτ), where A is the hyperfine coupling of a given nucleus. As a result, the signals vanish (give ‘blind spots’) at, Aτ = n, n = 0, 1, …, and show maximum intensities at Aτ = n+1/2. However, the chief use of pulsed ENDOR in this work was to observe very weakly coupled 2H signals, so that this suppression effect is of little consequence. Pulsed ENDOR measurements were implemented with random hopping of the radio frequency over the frequency range for a spectrum, a procedure that improves intensity and signal shape.
The ENDOR pattern for a single orientation of an I = 1/2 nucleus with relatively small hyperfine coupling and large gN, as is the case for 1H, exhibits a ν(±) doublet that is centered at the nuclear Larmor frequency, νN, and split by half the hyperfine coupling, |A/2|. For an I = 1 nucleus, such as 2H, there is quadrupole splitting such that each hyperfine-split doublet is further split into 2I lines. However, if the hyperfine and quadrupole coupling are of similar magnitude, then this first order pattern is not obtained. For typical conditions used here: g ~ 2.0, νmw ~ 35 GHz, B0 = 1.25 T, so that ν(1H) ≈ 53 MHz and ν(2H)) ≈ 8.2 MHz).
High Frequency (130 GHz, “D-band”) EPR and ENDOR spectra were obtained on a spectrometer described previously (41, 42). Field swept two-pulse (Hahn) echo-detected spectra were registered with 0/180° phase cycling of the first microwave pulse to eliminate baseline artifacts. Specific acquisition parameters are given in the figure legend.
Kinetic assay
To follow the slow kinetics related to radical decay, a diode array spectrophotometer (model DT 1000A, Analytical Instrument Systems, Inc., Flemington, NJ) was used. For observation of the rapid reaction related to radical formation, stopped-flow experiments were carried out on an Applied Photophysics spectrophotometer (SX.MV18, Leatherhead, U.K.) equipped with a photodiode array detector. The stopped-flow instrument was located in a Vacuum Atmospheres anaerobic chamber. In these mixing experiments, one syringe contained a solution consisting of 30 μM MCRred1 and CoBSH (or CoB6SH), and the other contained BES. The concentrations of CoBSH (or CoB6SH) and BES were varied in independent experiments. All experiments were performed in 50 mM Tris-HCl buffer, pH 7.6, at room temperature in the anaerobic chamber maintained at <0.5 ppm O2. Data were globally fit using the software pro-kineticist v 1.06 (Applied Photophysics Ltd., Leatherhead, U.K.).
RESULTS
Optical Spectroscopy
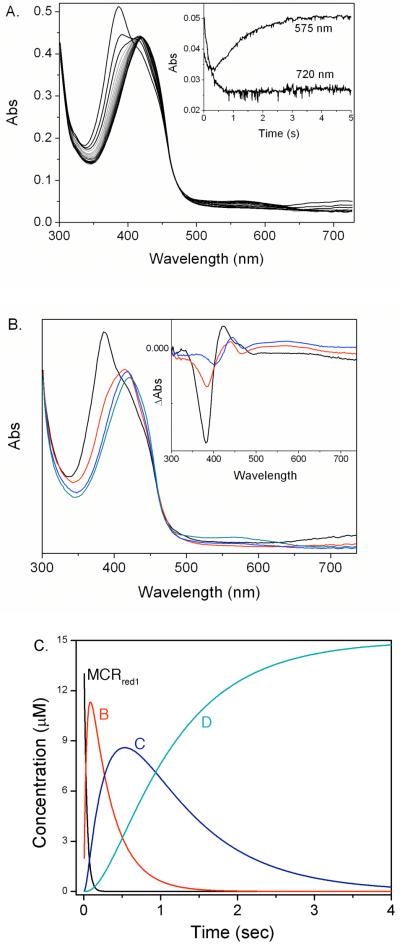
Upon addition of BES to MCRred1 in the presence of CoB6SH, the absorption maximum of MCRred1 (Fig. 1, solid line) at 385 nm shifts to 426 nm. Furthermore, the Ni(I)-associated peak at 720 nm disappears while a broad peak around 575 nm appears (dashed line), as clearly shown by the difference spectrum given in the inset. As described below, these absorption changes coincide with the formation of an EPR-active species that we will call “MCRBES”, or the “MCRBES radical”. The maximum difference absorption peaks associated with formation of the MCRBES radical include a rather sharp peak at 442 nm and a broader band around 575 nm. The addition of CoB6SH alone has no effect on the UV-visible spectrum of MCRred1, while the addition of BES alone results in decay of the 385 nm and 720 nm bands of MCRred1 without the appearance of the absorption feature at 575 nm.
Figure 1.
Visible spectra of the MCRBES radical. Visible spectrum of 21 μM MCRred1 (61%) (solid line) in the presence of 500 μM CoB6SH 30 s (dashed line) and 1 h (dotted line) after addition of 2 mM BES. The inset is the difference spectrum (the 30 s spectrum minus the spectrum before addition of BES).
The radical species generated by the reaction of MCRred1 in the presence of CoB6SH with BES is relatively stable; after one hour, the peak at 426 nm slightly decreases as the 575 nm peak disappears (Fig. 1, dotted line). The rates at which the absorption bands at 426 nm and 575 nm decrease were measured by fitting the data to a single-exponential decay function (Fig. 2), revealing a decay rate constant of 0.0021 ± 0.0003 s−1. When the solution containing MCRred1 and the native substrate CoBSH was reacted with BES, the decay rate constant was ~10-fold slower (0.00020 ± 0.00008 s−1, see Fig S1, Supporting Information). Thus, the presence of the native substrate CoBSH stabilizes the MCRBES radical (relative to CoB6SH).
Figure 2.
Slow kinetics of MCRBES radical decay after reaction of MCRred1 with BES in the presence of CoB6SH followed by visible (left) and EPR spectroscopy (squares). The EPR signal intensity of the MCRBES radical was measured after freeze quenching the reaction at various times. EPR parameters: microwave frequency, 9.384 GHz; microwave power, 10.440 mW; modulation frequency, 100 kHz; modulation amplitude, 10 G and temperature, 70 K. The calculated values were: kobs of 0.0021 ± 0.003 s−1 (UV-visible) or 0.0021 ± 0.0003 s−1 (EPR).
EPR Spectroscopy
Addition of BES to a solution of MCRred1, which is characterized by g-values at g⊥ = 2.065, g∥ = 2.24 (Fig. 3A), containing CoB6SH led to decay of the MCRred1 X-band EPR signal (measured at 70 K) coupled to the generation of a radical signal centered at g ≈ 2.002 with a doublet splitting of approximately 17 G (Fig. 3B); a simulation of this signal using an isotropic 1H hyperfine coupling of 50 MHz is also included. The peaks with values of g⊥ = 2.16 and g∥ = 2.23 derive from the MCRox1 state, which exists in variable proportions in MCR samples and has been assigned to a Ni(III)-thiolate species in resonance with a Ni(II)-thiyl radical (43, 44). There are small features at a g value of 2.07, which is characteristic of MCRred2 (45), and at g ~2.14 (of unknown origin) that are more easily resolved in the 35 GHz EPR spectrum (Figure S1, Supporting Information), but are not related to the scope of this investigation on the MCRBES radical. As shown in Figure 3, the EPR signal from MCRox1 is not affected by the reaction with CoB6SH and BES, indicating that only the Ni(I) state of MCR reacts with BES in the presence of CoB6SH. This MCRBES radical signal is also observed when MCRred1 is reacted with BES in the presence of the native substrate, CoBSH (data not shown). As with the UV-visible experiments, both BES and CoBSH (or CoB6SH) are required for radical formation; no changes in the MCRred1 EPR spectrum are observed when CoB6SH or CoBSH alone is added, and when only BES is added, the Ni(I) state of MCR oxidizes to EPR-silent Ni(II).
Figure 3.
EPR spectra of 150 μM MCRred1 (A) after addition of 1.5 mM CoB6SH and 1.5 mM BES in H2O (B) and D2O (C). The EPR spectrum in (D) was obtained after reacting MCRred1 containing universally labeled 2H6-Tyr residues under identical conditions as in (B). EPR parameters: microwave frequency, 9.384 GHz; microwave power, 10.440 mW; modulation frequency, 100 kHz; modulation amplitude, 10 G and temperature, 70 K.
To test the hypothesis that formation of the MCRBES EPR signal is coupled to the UV-visible spectra with difference peaks at 442 nm and 575 nm, freeze-quench EPR experiments were performed under conditions similar to the stopped flow experiments described above (Fig. 2). Thus, when a solution of MCRred1 and CoB6SH was reacted with BES, the EPR signal of the MCRBES radical decayed with the same rate constant (0.0021 ± 0.003 s−1) (Fig. 2, squares) that was measured by UV-visible spectroscopy (above, 0.0021 s−1). These experiments indicate that the 575 nm peak observed in the UV-visible spectrum and the MCRBES radical EPR spectrum originate from the same species.
To determine if the MCRBES radical contains exchangeable protons, the reaction of MCRred1 with CoB6SH and BES was carried out in both H2O and D2O and the resultant solutions examined by EPR and ENDOR spectroscopic experiments. As shown in Fig. 3C, the 1H doublet splitting in the X-band EPR spectrum of the radical is unchanged when the reaction is performed in D2O, demonstrating that the hyperfine splitting of the MCRBES radical is not associated with a solvent-exchangeable hydrogen nucleus.
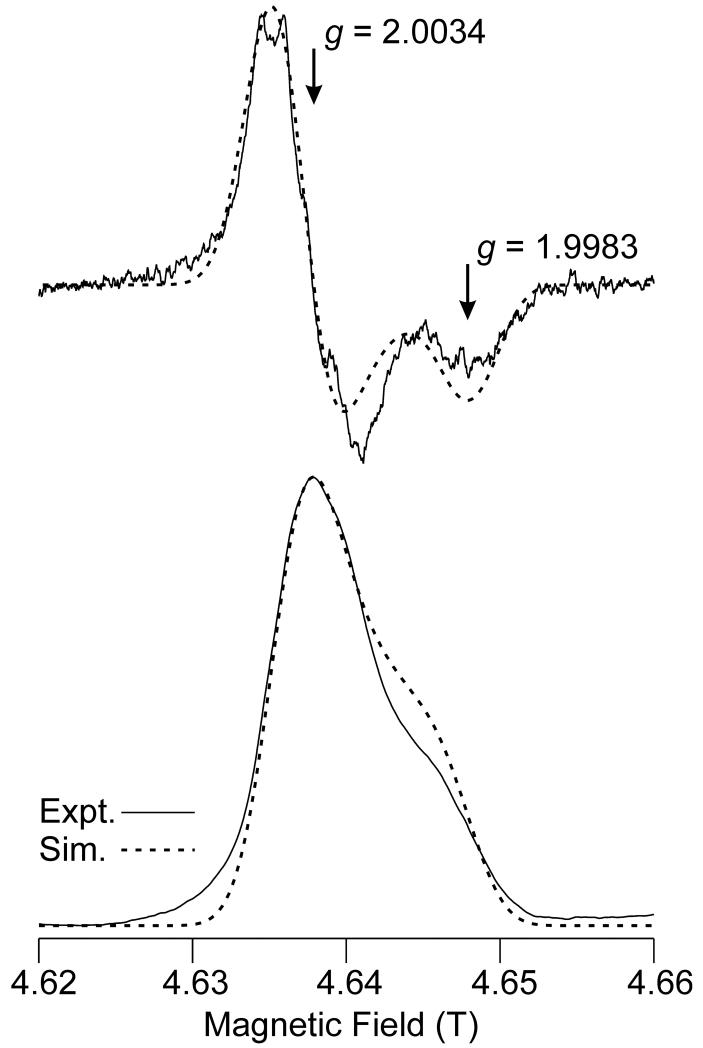
Accurate assignment of the g-values in the EPR spectra of organic radicals can aid in the identification of the radical. To obtain highly accurate g-values for the MCRBES radical, we recorded high frequency (HF) EPR spectra at 135 GHz (D-band). This high microwave frequency has the advantage of yielding better resolution of multiple EPR-active species in a heterogeneous sample (i.e., by providing greater dispersion of g values) and of revealing features that permit determination of the full g tensor of seemingly isotropic radical centers. Although the MCRBES radical appeared as a doublet with an isotropic g = 2.00, in the X-band EPR spectra, the Hahn echo-detected 135 GHz EPR spectrum displays approximate axial symmetry and lies entirely below g = 2.005 (Fig. 4). Although the majority of the spectrum appears to originate from a single radical species, it is possible that a small amount (<10%) of a minority species is present. Structure is evident in Fig. 4, but its origin is not known at this point and no attempt was made to simulate it.
Figure 4.
High Frequency (130 GHz) two-pulse (Hahn) echo-detected EPR spectrum of MCRred1 reacted with CoB6SH and BES for 1 min and then frozen. The derivative of the spectrum is also presented (upper trace) to approximate the effects of field modulation in a CW experiment. Acquisition parameters: temperature: 7 K; repetition rate: 30 Hz; number of acquisitions per point: 840; 90 degree pulse width: 40 ns; time between pulses: 150 ns; scan time: 200 s; number of scans: 28, EPR frequency: 130.001 GHz. Simulation parameters: g = [2.00340, 2.00340, 1.99832]; single-crystal Gaussian linewidths (hwhm), W = 60, 60, 50 MHz; isotropic hyperfine coupling to a single 1H with A = 50 MHz was also included, but this has a barely perceptible effect on the spectral appearance.
Simulation of the D-band EPR spectrum yielded g⊥ = 2.0034 and g∥ = 1.9983. This ability to accurately determine the g tensor of the MCRBES EPR signal in the HF spectra comes at the cost of increased linewidth due to g-strain (46). In the present case, the single-crystal linewidth increases from 27 MHz at X-band to 60 MHz at D-band, so that the 17 G splitting evident in the X-band spectra (Fig. 1A) is no longer resolved (simulations of the D-band spectrum were almost identical whether or not an isotropic 1H hyperfine splitting of 50 MHz, as determined at X-band, was included). Likewise, the resolved 14N hyperfine splitting at g⊥ of the MCRred1 signal (from the F430 pyrrole/pyrroline ligands (43, 45)) evident in the X-band spectrum is not resolved in the 135 GHz or the 35 GHz spectra (see Fig. S2, Supporting Information).
Q-Band (35 GHz) and D-Band (135 GHz) ENDOR spectroscopic experiments
The 35 GHz ENDOR methodology employed here uses a temperature of 2 K, which is best suited to metal-centered paramagnets. Thus, it was comparatively easy to record good quality spectra on field positions of the MCR Ni signals (e.g., g ~ 2.17), which readily showed the ν+ partner of the strongly coupled (A(1H) = 28 MHz) signal that has been attributed to a hydrido ligand (denoted Hrh) to Ni(III) in MCRred2a (47). This signal can be seen in Fig. 5. In this case, CW ENDOR with random hopping of the rf was employed, which gives a better baseline. A corresponding swept ENDOR spectrum is shown in Figure S3 (Supporting Information). In contrast, spectra recorded at the field position of the radical signal (g ~ 2.00) were of lower signal/noise ratio and did not exhibit any strongly coupled protons; only those with A(1H) < ~12 MHz were observed, as also seen in Fig. 5. Swept rf CW 1H Q-band ENDOR spectra recorded at g ~2.00 (radical signal) and at g ~ 2.17 (MCR signal) are shown in Fig. S3. Under these conditions, the 1H ENDOR signal has a higher signal/noise ratio. There appear to be signals extending to > 20 MHz from ν(1H), so that the maximum hyperfine coupling may be > 40 MHz, in agreement with the X-band EPR results. Note that at 35 GHz, the radical EPR signal still appears isotropic (in contrast to the 130 GHz result, Fig. 4). As a result, there is no “orientation selection” in the ENDOR, in contrast to the anisotropic MCR signal(s), which leads to broader ENDOR lines and greater difficulty in determining the A(1H) tensor. No significant differences were seen in the 1H pattern for H2O versus D2O samples.
Figure 5.
Broad scan “stochastic” CW 35 GHz 1H ENDOR spectra recorded at 2 K of MCRred1 in the presence of 1 mM CoB6SH after addition of BES at a field position on the radical signal (blue trace; g = 2.008) and at one on the MCR signal (red trace; g = 2.175). The latter exhibits a signal from a strongly coupled 1H that has been denoted as Hrh by Harmer et al. and fully analyzed by them.(47) Experimental conditions: microwave frequency, 35.013 GHz; modulation frequency, 1 G; microwave power, 30 dB (~10 μW); random hopping of rf with delay time: 1 ms, sample time: 1 ms, rf time: 1 ms; 400 scans. The inset shows 35 GHz 2H Mims ENDOR spectra at 2 K of MCRred1 in H2O solvent (blue trace) and D2O solvent (green trace). The spectra are centered at the 2H Larmor frequency (8.1 MHz) with a scale in 2H coupling, but a scale in corresponding 1H coupling is also shown. Experimental conditions: microwave frequency, 34.895 GHz (D2O), 34.851 GHz (H2O); π/2 pulse width = 50 ns; delay time, τ = 600 ns; magnetic field, 1.2418 T (g = 2.0077 (D2O); 2.0052 (H2O)); repetition time, 20 ms; random hopping of rf; 50 scans.
The use of 2H ENDOR facilitates focusing on the question of solvent exchange more directly. Fig. 5 (inset) compares the 35 GHz Mims 2H ENDOR spectra of the MCRBES samples generated in H2O and D2O, recorded at the field position corresponding to the maximum intensity of the radical signal. The sample in D2O showed a largely unresolved matrix 2H ENDOR signal centered at the 2H Larmor frequency that is essentially the same when recorded at varying field positions on the EPR envelope of the radical signal. Superimposed on this is a poorly resolved splitting, which might be assigned to A(2H) ≈ 0.3 MHz (corresponding to A(1H) ≈ 2 MHz, which is well inside the overall 1H CW ENDOR pattern, Fig. S1).1 In summary, the combined ENDOR and EPR results rule out the possibility that the doublet splitting seen in the X-band EPR spectrum derives from an exchangeable proton.
Isotopic substitution and EPR/ENDOR spectroscopic experiments to determine if the MCRBES species derives from a tyrosyl radical
Based on the crystal structure of MCR (3), there are a number of viable candidates for the relatively stable MCRBES radical. These include: αTyr333 (4.3 Å from Ni), βTyr367 (4.3 Å from Ni), αthioglycine445 (12 Å from Ni), the sulfur of CoBSH (8.87 Å away from Ni) (6) or the corphin ring itself. Given how close αTyr333 and βTyr367 are to the Ni active site, they seemed to be the most likely contenders.
Tyrosyl radicals are widely found in enzymes, including the photosynthetic oxygen evolving complex (OEC) (48, 49), prostaglandin H synthase (PGHS) (50, 51), ribonucleotide reductase (RNR) (52) and galactose oxidase (53, 54). The low frequency (X-band) EPR spectra of tyrosyl radicals are dominated by hyperfine coupling to non-exchangeable protons. Each of the two ring protons ortho to the phenol oxygen have an anisotropic coupling, the largest component of which is typically in the 20 to 30 MHz range. The two side-chain methylene protons have an approximately isotropic coupling, the values of which are highly dependent on the dihedral angle with respect to the ring plane normal. Depending on the angle, the coupling of these protons may range from ~ 0 to > 60 MHz. However, no matter what the angle, at least one methylene proton will experience a significant (>10 MHz) hyperfine coupling. The variability of the side chain dihedral angle, and resulting variability of the methylene proton hyperfine couplings, means that the X-band spectra of tyrosyl radicals can display a variety of splitting patterns. But all tyrosyl radical spectra experience significant spectral splittings and/or width from the hyperfine coupling of 3 or 4 non-exchangeable protons, and thus the substitution of natural abundance tyrosine by an isotopolog that is deuterated at any position would be expected to have an observable spectral impact for any tyrosyl radical species.
Unlike X-band spectra, the D-band spectra of tyrosyl radicals are dominated by the (field-dependent) Zeeman interaction, which is gauged by g-values. Unmodified tyrosyl radicals have an anisotropic (rhombic) g-matrix with principal values that are fairly consistent: gx (or g1) is in the range of 2.009 −2.006 depending on the electrostatic/hydrogen bonding environment of the phenol oxygen; gy (or g2) ~2.004 - 2.005; and gz (or g3) ~ 2.002 - 2.003. Thus, by using HFEPR to determine the g-values, as well as X-band EPR and Q-band ENDOR to measure the hyperfine couplings of the radical in the enzyme prepared with both protonated and deuterated tyrosine, we sought to provide definitive evidence for or against the radical species originating from tyrosine, specifically αTyr333 or βTyr367.
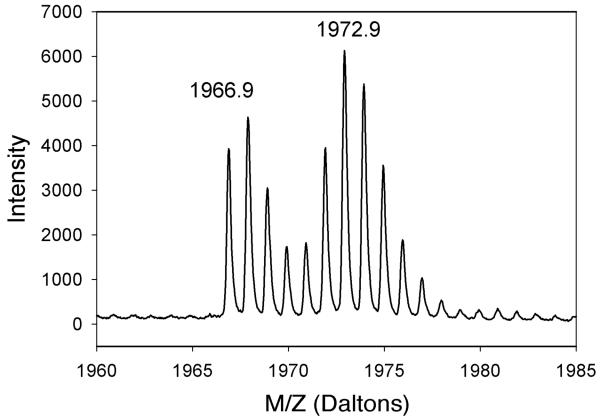
Samples of MCR were generated that contained a tyrosine isotopolog. MCRred1 was globally labeled with 2H6-Tyr by feeding M. marburgensis cells with 2H6-hydroxyphenylacetate; then MCR was isolated and digested, and the amino acids were analyzed with GC-MS. The results showed that at least 50% of tyrosine residues in MCR were labeled with 2H4-Tyr instead of 2H6-Tyr. This is a lower estimate of the labeling percentage because significant H-exchange occurs during the acid hydrolysis. A similar estimate of the labeling percentage was obtained by mass spectrometric analysis of BrCN- and AspN-digested fragments of MCR isolated from cells grown on labeled hydroxyphenylacetate, e.g., greater than 50% of βTyr367 was labeled (Fig. 6). Even though the fragment including αTyr333 was not identified, eleven other peptides containing tyrosine residues showed a similar percentage of 2H6-Tyr labeling. Thus, independent amino acid and peptide analyses confirmed that the Tyr residues in MCR were significantly labeled when cells were fed with 2H6-hydroxyphenylacetate under the conditions described.
Figure 6.
Mass spectrum of the fragment containing βTyr367 (IYGGGGPGIFNGNHIVTRH), which is located directly above the Ni center in F430. MCR isolated from M. marburgensis cells was digested with BrCN and AspN.
In order to test the hypothesis that the radical intermediate detected by EPR and UV-visible spectroscopic studies derives from Tyr, 2H6-Tyr-labeled MCRred1 was reacted with BES in the presence of CoB6SH. If the EPR spectra were associated with one of the two Tyr residues located immediately above the Ni center at the active site of MCR, then a significant change in the lineshape would be expected upon substitution of these 1H6-Tyr residues with 2H6-Tyr. As shown in Fig 3D, the EPR spectrum of the MCRBES radical appeared unchanged by the 2H-Tyr substitution, with the same degree of hyperfine splitting remaining in the MCRBES radical. These experiments unambiguously rule out the possibility that MCRBES contains a tyrosyl radical.
Pulsed 35 GHz ENDOR spectra of the sample containing 2H6-Tyr exhibited a very weak and featureless signal centered at the deuteron Larmor frequency. Essentially the same signal (not shown) was observed at multiple positions on the EPR envelope. This 2H signal may arise from distant coupling to the various paramagnetic centers that make up the overall EPR envelope. The 1H ENDOR spectrum of this sample was indistinguishable from the samples with natural abundance tyrosine whether in H2O or D2O solvent.
Effect of tyrosyl acetylation
Even though the EPR experiments clearly show that MCRBES radical is not a tyrosyl radical, the tyrosyl residues still appear to play an important role in radical formation. As shown in Fig 7A, acetylation of tyrosyl residues in MCRred1 by treatment with N-acetylimidazole has no effect on the EPR spectrum of MCRred1; however, Tyr acetylation blocks the formation of the MCRBES radical. As shown in Fig 7C, the amount of MCRBES generated from the reaction of acetylated MCRred1 with BES and CoB6SH was greatly decreased and some MCRred1 remains, as compared with the reaction of unlabeled MCRred1 with BES and CoB6SH (Fig. 7B).
Figure 7.

X-band EPR spectra of 100 μM MCRred1 after reaction with (A) acetyl-imidazole, (B) with 5 mM BES in presence of 1 mM CoB6SH, and (C) with 5 mM BES and 1 mM CoB6SH (as in B) and acetyl-imidazole. EPR parameters: microwave frequency, 9.384 GHz; microwave power, 10.440 mW; modulation frequency, 100 kHz; modulation amplitude, 10 G and temperature, 70 K.
Kinetics of MCRBES radical formation
The MCRBES radical is formed rapidly and the transient kinetics were followed by stopped-flow with UV-visible spectroscopy (Fig 8A). The kinetic data ranging from 350 nm to 732 nm were globally fit to a four-component equation (eq 2). Two spectral intermediates, B (Fig 8B, red line),
| eq 2 |
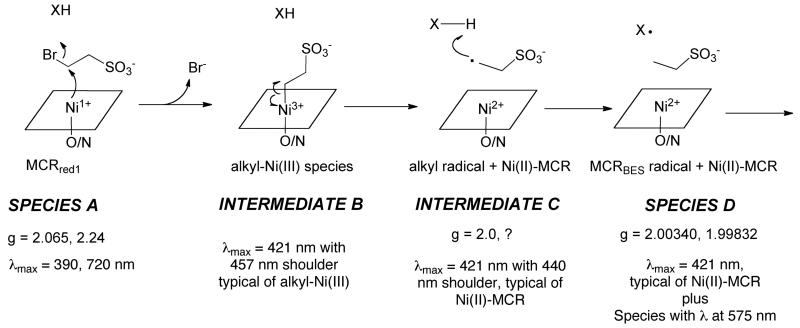
C (Fig 8B, blue line) and D (associated with the MCRBES radical) were identified. The difference spectra between A and B (Fig 8B inset, black line) showed that a new species with absorption at 420 nm and a shoulder at 457 nm was formed accompanying the decay of the 385 and 720 nm bands of MCRred1. It has been shown that the shoulder around 457 nm is characteristic of the methyl-Ni(III) species of MCR (14); thus, we assign intermediate B as an alkyl-Ni(III) species. The 457 nm peak then disappears as intermediate B converts to intermediate C, which appears to be a Ni(II) species. In the last step, the MCRBES radical (Species D) forms.
Figure 8.
(A) Time-course of the reaction of 15 μM MCRred1 with 5 mM BES in the presence of 110 μM CoB6SH (interval 0.1 s). The Insert shows the absorbance changes at 575 and 720 nm. (B) Visible spectra of MCRred1 plus CoB6SH (black line), the trapped intermediate B (red line), the trapped intermediate C (blue line) and Intermediate D (including the MCRBES radical, green line) by global fitting the data in (A) to four-component reaction. Insert: The difference spectra of intermediate B minus MCRred1 (black line), intermediate B minus intermediate C (red line) and MCRred1 minus intermediate C (blue line). (C) The time-course of the changes in the concentrations of MCRred1 (black line), intermediate B (red line), intermediate C (blue line) and Intermediate D, the MCRBES radical (green line) upon the reaction in (A).
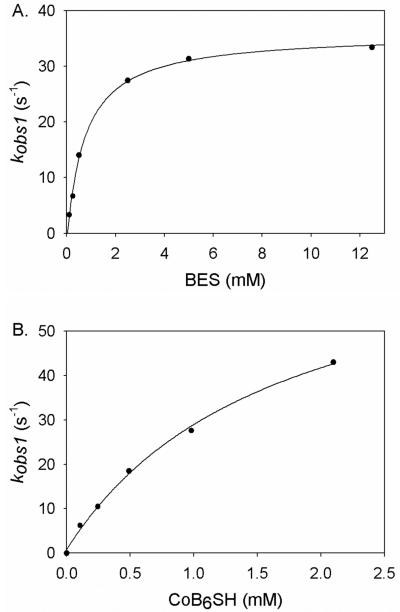
By varying the concentration of BES (Fig 9A) or CoB6SH (Fig 9B), the first step in this three-step sequence (with rate constant kobs1) was shown to be dependent on the concentrations of both CoB6SH and BES, while kobs2 and kobs3 were independent of the CoB6SH or BES concentrations. The kobs1 was fit to equation 3 to derive the values of k1 and Km, where [S] is the concentration of CoB6SH or BES.
| eq 3 |
The rate constants for the three steps are provided in Table 1. The conversion of A to intermediate B gives a second order rate constant of 45,000 M−1s−1, the conversion of intermediate B to C has a rate constant of 2.6 s−1 and formation of the MCRBES radical (species D) occurs with a rate constant of 1.0 s−1. As shown above, decay of Species D occurs with a rate constant of 0.0021 s−1 (not given in Table I).
Figure 9.
Concentration dependence of the rate of MCRBES radical formation (A) Dependence of MCRBES radical formation on the concentration of BES. A solution containing 15 μM MCRred1 (58%) and 1 mM CoB6SH was reacted with varied concentrations of BES at room temperature. (B). CoB6SH concentration dependence of MCRBES radical formation. The mixture of 15 μM MCRred1 (58%) and various concentration of CoB6SH was shot against 5 mM BES at room temperature. Reported rate constants are the average of three to four different rapid-mixing experiments.
Table 1. Kinetic parameters for the three-step reaction involving MCRred1 radical formation.
| k1 (s−1) | Km1 (mM) | k1/Km1 (M−1s−1) | k2 (s−1) | k3 (s−1) | |
|---|---|---|---|---|---|
| CoB6SH (with 5 mM BES) | 75 ± 6 | 1.7 ± 0.3 | 45000 ± 9000 | 2.6 ± 0.3 | 1.0 ± 0.1 |
| BES (in presence of 1 mM CoB6SH) | 38 ± 1 | 0.7 ± 0.1 | 51500 ± 9900 | 2.8 ± 0.5 | 1.1 ± 0.1 |
DISCUSSION
Transient kinetics combined with UV-visible and EPR spectroscopic experiments have revealed the accumulation of three intermediates during the reaction of the active Ni(I)-MCRred1 with the substrate analogs CoB6SH and BES (Figure 8). Figure 10 describes the mechanism by which the various species form. The first of these intermediates is assigned as an alkyl-Ni(III) species because it exhibits an absorption spectrum similar to that of the organonickel intermediates formed when MCRred1 reacts with various alkyl halides including BPS and methyl iodide (14, 17). When reacted with thiolates (including HSCoM) (12, 16, 28), these alkyl-Ni(III) species convert to the active MCRred1 state along with the corresponding alkane when protonolyzed or reduced with Ti(III) citrate (14, 17), supporting the catalytic intermediacy of the alkyl-Ni species in methane synthesis. Prior studies indicated that reactive haloalkyl compounds that are the size of HSCoM generate radicals, while those that are the size of methyl-SCoM (and larger) yield organometallic species. However, as shown here, the reaction with BES also generates an organometallic intermediate that rapidly converts to the characteristic MCRBES radical. Therefore, the reaction of MCR with all haloalkanes may generate an initial organometallic species that converts to other intermediates at varying rates that determine whether or not these species accumulate to detectible levels. This organonickel species (B) quickly converts to a relatively stable radical labeled as X· in Figure 8 (Species D, MCRBES) through Intermediate C, which was identified by global kinetic analysis. We propose that MCRBES is formed by homolytic cleavage of the alkyl-Ni(III) bond to generate an unstable alkyl radical that would abstract a hydrogen atom from the precursor of X· (i.e., X-H).
Figure 10.
Proposed mechanism for MCRBES radical formation. “X” is the species with the doublet radical EPR spectrum and the long wavelength absorption. Further isotope substitution and spectroscopic experiments are required to identify “X”, as described in the text.
Free radicals have been identified as intermediates in a number of enzymatic reactions (55). These radicals include amino acid-based radicals, such as the tyrosyl radical in class I ribonucleotide reductases (52) and in other enzymes (49, 51, 53), a cysteine-based thiyl radical in all three classes of ribonucleotide reductase (55), and a glycyl radical in pyruvate formate lyase (56). Furthermore, catalytic radicals are formed by various cofactors, e.g., the adenosyl radical (generated from S-adenosyl-L-methionine or adenosylcobalamin), a hydroxyethyl thiamine pyrophosphate radical in pyruvate ferredoxin oxidoreductase, and a porphyrin π-cation radical in chloroperoxidase (55, 57, 58).
Radicals have been proposed as key intermediates in the MCR-catalyzed synthesis of methane (6, 22, 23); however, so far such radicals have not been identified spectroscopically when MCR is reacted with its natural substrates. On the other hand, a radical EPR doublet signal (the MCRBES radical) is generated when MCRred1 is reacted with BES in the presence of CoB6SH/CoBSH (18). We show here that MCRBES is transiently formed and accumulates to a maximum of ~20 % of the initial amount of MCRred1.
Based on the crystal structure of MCR (3), it appears that several amino acids (Tyr and thioglycine), the corphin ring itself, and the sulfur group of its substrates, CoB6SH/CoBSH or HSCoM, are reasonable candidates for XH, the precursor of the MCRBES radical. Although the simplest explanation for the 575 nm absorption peak and the MCRBES radical to accumulate over the same time frame is that they belong to the same species, another possibility is that the EPR and UV-visible spectra belong to different, but kinetically coupled, species. This is because transient kinetic experiments assign intermediates based on the dynamics of spectral evolution. For example, homolytic cleavage of alkyl-Ni(III) would generate the alkyl radical and a Ni(II) species, which would be assigned as a single intermediate by the global kinetic fitting protocol, because they form and decay at the same rate. According to this scenario, the UV-visible spectrum would be dominated by the Ni(II) component and the EPR spectrum would reveal the radical. As described above, because two Tyr residues are situated just above the upper axial Ni ligand (the ethyl sulfonyl group in this case), we used EPR and ENDOR spectroscopy to test the possibility that the MCRBES radical comes from Tyr.
Our EPR and ENDOR spectroscopic studies of the MCRBES radical generated from 2H6-Tyr-labeled MCR rule out the possibility that the MCRBES radical derives from tyrosine. First, the g-values of the MCRBES radical (g⊥ = 2.0034 and g∥ = 1.9983) are not consistent with those typically observed for a Tyr radical which exhibits rhombic spectra with g1 is in the range of 2.009 to 2.006, g2 at 2.004 - 2.005 and g3 at 2.002 - 2.003. The g-values of the MCRBES radical are most consistent with a species with significant electron spin density localization on carbon, hydrogen and/or nitrogen rather than oxygen and sulfur, since the latter would be expected to result in g values > 2.005 (for sulfur-based radicals, see (59-61)). Furthermore, as described in the Results section, because all tyrosyl radical spectra exhibit significant broadening or splitting from the hyperfine interactions of 3 or 4 non-exchangeable protons, the substitution of natural abundance Tyr by 2H-Tyr would be expected to markedly alter the spectrum of any tyrosyl radical species. The lack of any observable difference in the EPR spectra of the MCRBES radical generated from enzyme containing 2H- versus 1H-Tyr demonstrates conclusively that MCRBES is not a Tyr radical.
The visible spectrum, with a broad peak around 575 nm, is also inconsistent with a typical Tyr radical. For example, the primary absorption bands characteristic for a phenoxyl radical include a sharp intense band at ~410 nm (ε > 1800 M−1cm−1), another intense but broader band at ~390 nm (ε > 900 M−1cm−1) and a broad weak band at ~600 nm (ε < 500 M−1cm−1) (62, 63). Although the absorption spectrum is not consistent with a Tyr radical, it would not be inconsistent with that of a radical formed on the corphin ring of F430. A porphyrin π-radical, which is coupled with an oxoiron(IV) heme species, has been reported in many hemoproteins (57) and has a characteristic absorbance in the range of 650 to 700 nm (64). Because the corphin ring contains only five double bonds, four of which are in conjugation, the absorption maxima of a corphin-based radical could shift to higher energies. However, as mentioned below, the EPR spectra are inconsistent with assigning the radical to the tetrapyrrole ring of F430. Alternatively, as mentioned above, one can consider the possibility that the EPR and UV-visible spectra belong to different, but kinetically coupled, species.
Glycyl, tryptophanyl, ketyl, allyl and other C/N based radicals as well as the corphin-based radical may have g values in the range observed here, but these species have hyperfine couplings that are either not fully characterized or are inconsistent with those measured here. For example, although it is an otherwise attractive possibility given the long wavelength absorption (see below), it appears highly unlikely that the MCRBES radical derives from the corphin ring of F430 because the Ni(II) center in MCR and in the free cofactor has an S = 1 configuration with a significant zero field splitting (65). Thus, such a radical would likely magnetically interact with the tetrapyrrole in a way that would lead to a significantly different EPR spectrum that that observed. Rather than being a narrow EPR signal at g ≈ 2.00, a coupled radical would have substantial g anisotropy. It appears that the observed EPR spectrum could only arise from a tetrapyrrole-centered radical if the Ni(II) center were in the S = 0 configuration, which has no precedence in MCR forms. Thus, although a Tyr radical can be ruled out, further experiments, including isotopic substitutions, will be required to make a positive assignment for the MCRBES radical.
Even though MCRBES (X· in Figure 10) does not contain a tyrosyl radical, our results suggest that a tyrosyl residue may still play a role in formation of the MCRBES radical. Acetylation of tyrosyl residues greatly inhibits the reaction of MCRred1 with BES, suggesting that a tyrosyl residue may be involved in formation of MCRBES radical. The acetyl group may simply sterically hinder the reaction of Ni(I) with BES and MCRBES may be formed by a pathway independent of the involvement of Tyr. However, another hypothesis that is consistent with our Tyr-modification results is that the stable doublet MCRBES radical with the long wavelength absorbance is generated by hydrogen atom abstraction from a transient Tyr radical. According to this scenario, the ethanesulfonyl radical (Intermediate C) could abstract a hydrogen atom from one of the two nearby Tyr residues to form a tyrosyl radical that could rapidly abstract a hydrogen atom from the corphin ring or one of the other species described above (CoBSH/CoB6SH, thiogycine, HSCoM) to generate the MCRBES radical (Species D). In this scenario, the Tyr radical would form and decay too rapidly to accumulate to detectable levels. The intermediacy of the Tyr radical is, of course, speculative since, thus far, the only radical we have observed is the relatively stable MCRBES radical with the doublet EPR spectrum.
The detection of both organonickel and radical species suggests a new mechanism for methane synthesis that combines the key features of Mechanisms I (a methylnickel intermediate) and II (a methyl radical intermediate). Both mechanisms also predict that a thiyl-radical from CoBSH is generated as an intermediate, but the EPR parameters of the MCRBES radical do not support a thiyl radical assignment for the doublet EPR signal. Because of the inhibitory effects of Tyr acetylation, we speculate that the stable radical is formed by abstraction of a H-atom from an initially formed Tyr radical. The final proposed steps in both Mechanisms I and II involve a disulfide anion radical (CoBS-SCoM) that reduces the Ni(II) species (above) back to active MCRred1. Our studies also provide evidence against a potential F430-based radical, which was invoked in Mechanism III.
Because no intermediates have been observed in various rapid kinetic studies of the reaction of MCRred1 with the native substrates, it is important to identify the MCRBES radical by performing further isotope labeling studies. Recent experiments of the reaction of MCRred1 with methyl-SCoM and CoB6SH reveal a doublet radical, but with different UV-visible spectra than those of MCRBES (unpublished, Dey & Ragsdale). Thus, experiments similar to those described here may reveal the identity of an intermediate in the reaction with methyl-SCoM and CoBSH.
Supporting Informational material is available free of charge via the Internet at http://pubs.acs.org and contains the following: Kinetics of MCRBES radical decay with BES and CoBSH as substrates (Figure S1), 35 GHz EPR spectra at 2 K of MCRred1 in presence 1 mM CoB6SH after addition of BES (Figure S2), and Broad scan swept CW 35 GHz 1H ENDOR spectra of MCRred1 recorded at 2 K at a field position on the MCR signal (g = 2.177, upper trace) and on the radical signal (g = 2.002, lower trace) (Figure S3).
Supplementary Material
Footnotes
Deuterium signals also were not observed in the D-band (130 GHz) ENDOR spectra of samples of MCRred1 that had been exchanged with D2O and incubated with BES and CoB6SH (not shown).
This work was supported by DOE (ER15931, SWR), NSF (MCB-0316038, BMH), and NIH (GM-075920 to GJG).
BRIEFS (WORD Style “BH_Briefs”). Methyl-coenzyme M reductase forms a radical intermediate that has been characterized by spectroscopic and kinetic methods.
- MCR
- Methyl-coenzyme M reductase
- BES
- bromoethane sulfonate
- BPS
- 3-bromopropanesulfonate
- DFT
- density functional theory
- ENDOR
- electron nuclear double resonance
- EPR
- electron paramagnetic resonance
- HF
- high frequency
- HSCoM
- Coenzyme M
- CoBSH
- Coenzyme B
- rf
- radiofrequency
REFERENCES
- 1.DiMarco AA, Bobik TA, Wolfe RS. Unusual coenzymes of methanogenesis. Annu Rev Biochem. 1990;59:355–394. doi: 10.1146/annurev.bi.59.070190.002035. [DOI] [PubMed] [Google Scholar]
- 2.Thauer RK. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. 1998 Marjory Stephenson Prize Lecture. Microbiology. 1998;144(Pt 9):2377–2406. doi: 10.1099/00221287-144-9-2377. [DOI] [PubMed] [Google Scholar]
- 3.Ermler U, Grabarse W, Shima S, Goubeaud M, Thauer RK. Crystal structure of methyl-coenzyme M reductase: the key enzyme of biological methane formation. Science. 1997;278:1457–1462. doi: 10.1126/science.278.5342.1457. [DOI] [PubMed] [Google Scholar]
- 4.Mahlert F, Bauer C, Jaun B, Thauer RK, Duin EC. The nickel enzyme methyl-coenzyme M reductase from methanogenic archaea: In vitro induction of the nickel-based MCR-ox EPR signals from MCR-red2. J Biol Inorg Chem. 2002;7:500–513. doi: 10.1007/s00775-001-0325-z. [DOI] [PubMed] [Google Scholar]
- 5.Grabarse W, Mahlert F, Shima S, Thauer RK, Ermler U. Comparison of three methyl-coenzyme M reductases from phylogenetically distant organisms: unusual amino acid modification, conservation and adaptation. J Mol Biol. 2000;303:329–344. doi: 10.1006/jmbi.2000.4136. [DOI] [PubMed] [Google Scholar]
- 6.Grabarse W, Mahlert F, Duin EC, Goubeaud M, Shima S, Thauer RK, Lamzin V, Ermler U. On the mechanism of biological methane formation: structural evidence for conformational changes in methyl-coenzyme M reductase upon substrate binding. J Mol Biol. 2001;309:315–330. doi: 10.1006/jmbi.2001.4647. [DOI] [PubMed] [Google Scholar]
- 7.Goubeaud M, Schreiner G, Thauer RK. Purified methyl-coenzyme-M reductase is activated when the enzyme-bound coenzyme F430 is reduced to the nickel(I) oxidation state by titanium(III) citrate. Eur J Biochem. 1997;243:110–114. doi: 10.1111/j.1432-1033.1997.00110.x. [DOI] [PubMed] [Google Scholar]
- 8.Albracht SPJ, Ankel-Fuchs D, Böcher R, Ellermann J, Moll J, van der Zwann JW, Thauer RK. Five new EPR signals assigned to nickel in methyl-coenzyme M reductase from Methanobacterium thermoautotrophicum, strain Marburg. Biochimica et Biophysica Acta. 1988;955:86–102. [Google Scholar]
- 9.Rospert S, Böcher R, Albracht SPJ, Thauer RK. Methyl-coenzyme M reductase preparations with high specific activity from H2-preincubated cells of Methanobacterium thermoautotrophicum. FEBS Lett. 1991;291:371–375. doi: 10.1016/0014-5793(91)81323-z. [DOI] [PubMed] [Google Scholar]
- 10.Goenrich M, Duin EC, Mahlert F, Thauer RK. Temperature dependence of methyl-coenzyme M reductase activity and of the formation of the methyl-coenzyme M reductase red2 state induced by coenzyme B. J Biol Inorg Chem. 2005;10:333–342. doi: 10.1007/s00775-005-0636-6. [DOI] [PubMed] [Google Scholar]
- 11.Ellermann J, Rospert S, Thauer RK, Bokranz M, Klein A, Voges M, Berkessel A. Methyl-coenzyme-M reductase from Methanobacterium thermoautotrophicum (strain Marburg). . Purity, activity and novel inhibitors. Eur. J. Biochem. 1989;184:63–68. doi: 10.1111/j.1432-1033.1989.tb14990.x. [DOI] [PubMed] [Google Scholar]
- 12.Rospert S, Voges M, Berkessel A, Albracht SP, Thauer RK. Substrate-analogue-induced changes in the nickel-EPR spectrum of active methyl-coenzyme-M reductase from Methanobacterium thermoautotrophicum. Eur J Biochem. 1992;210:101–107. doi: 10.1111/j.1432-1033.1992.tb17396.x. [DOI] [PubMed] [Google Scholar]
- 13.Hinderberger D, Piskorski RP, Goenrich M, Thauer RK, Schweiger A, Harmer J, Jaun B. A nickel-alkyl bond in an inactivated state of the enzyme catalyzing methane formation. Angew Chem Int Ed Engl. 2006;45:3602–3607. doi: 10.1002/anie.200600366. [DOI] [PubMed] [Google Scholar]
- 14.Dey M, Telser J, Kunz RC, Lees NS, Ragsdale SW, Hoffman BM. Biochemical and spectroscopic studies of the electronic structure and reactivity of a methyl-Ni species formed on methyl-coenzyme M reductase. J Am Chem Soc. 2007;129:11030–11032. doi: 10.1021/ja074556z. [DOI] [PubMed] [Google Scholar]
- 15.Yang N, Reiher M, Wang M, Harmer J, Duin EC. Formation of a nickel-methyl species in methyl-coenzyme M reductase, an enzyme catalyzing methane formation. J Am Chem Soc. 2007;129:11028–11029. doi: 10.1021/ja0734501. [DOI] [PubMed] [Google Scholar]
- 16.Dey M, Kunz RC, Lyons DM, Ragsdale SW. Characterization of alkyl-nickel adducts generated by reaction of methyl-coenzyme m reductase with brominated acids. Biochemistry. 2007;46:11969–11978. doi: 10.1021/bi700925n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunz RC, Horng YC, Ragsdale SW. Spectroscopic and kinetic studies of the reaction of bromopropanesulfonate with methyl-coenzyme M reductase. J Biol Chem. 2006;281:34663–34676. doi: 10.1074/jbc.M606715200. [DOI] [PubMed] [Google Scholar]
- 18.Goenrich M, Mahlert F, Duin EC, Bauer C, Jaun B, Thauer RK. Probing the reactivity of Ni in the active site of methyl-coenzyme M reductase with substrate analogues. J Biol Inorg Chem. 2004;9:691–705. doi: 10.1007/s00775-004-0552-1. [DOI] [PubMed] [Google Scholar]
- 19.Gunsalus RP, Romesser JA, Wolfe RS. Preparation of coenzyme M analogues and their activity in the methylcoenzyme M reductase system of Methanobacterium thermoautotrophicum. Biochemistry. 1978;17:2374–2377. doi: 10.1021/bi00605a019. [DOI] [PubMed] [Google Scholar]
- 20.Ellermann J, Hedderich R, Böcher R, Thauer RK. The final step in methane formation. Investigations with highly purified methyl-CoM reductase (component C) from Methanobacterium thermoautotrophicum (strain Marburg) Eur. J. Biochem. 1988;172:669–677. doi: 10.1111/j.1432-1033.1988.tb13941.x. [DOI] [PubMed] [Google Scholar]
- 21.Immig I, Demeyer D, Fiedler D, Van Nevel C, Mbanzamihigo L. Attempts to induce reductive acetogenesis into a sheep rumen. Arch. Tierernahr. 1996;49:363–370. doi: 10.1080/17450399609381898. [DOI] [PubMed] [Google Scholar]
- 22.Signor L, Knuppe C, Hug R, Schweizer B, Pfaltz A, Jaun B. Methane formation by reaction of a methyl thioether with a photo-excited nickel thiolate--a process mimicking methanogenesis in archaea. Chemistry. 2000;6:3508–3516. doi: 10.1002/1521-3765(20001002)6:19<3508::aid-chem3508>3.3.co;2-n. [DOI] [PubMed] [Google Scholar]
- 23.Pelmenschikov V, Blomberg MR, Siegbahn PE, Crabtree RH. A mechanism from quantum chemical studies for methane formation in methanogenesis. J Am Chem Soc. 2002;124:4039–4049. doi: 10.1021/ja011664r. [DOI] [PubMed] [Google Scholar]
- 24.Duin EC, McKee ML. A New Mechanism for Methane Production from Methyl-Coenzyme M Reductase As Derived from Density Functional Calculations. J Phys Chem B. 2008 doi: 10.1021/jp709860c. [DOI] [PubMed] [Google Scholar]
- 25.Lin S-K, Jaun B. Coenzyme F430 from Methanogenic bacteria: detection of a paramagnetic methylnickel(II) derivative of the pentamethyl easter by 2H-NMR spectroscopy. Helvetica Chimica Acta. 1991;74:1725–1738. [Google Scholar]
- 26.Lin S-K, Jaun B. Coenzyme F430 from Methanogenic bacteria: mechanistic studies on the reductive cleavage of sulonium ions catalyzed by F430 pentamethyl ester. Helvetica Chimica Acta. 1992;75:1478–1490. [Google Scholar]
- 27.Lahiri Goutam K, Schussel Leonard J, Stolzenberg AM. F430 model chemistry. Mechanistic investigation of the reduction, coupling, and dehydrohalogenation of alkyl halides by the nickel(I) octaethylisobacteriochlorin anion. Inorganic chemistry. 1992;31:4991–5000. [Google Scholar]
- 28.Kunz RC, Dey M, Ragsdale SW. Characterization of the thioether product formed from the thiolytic cleavage of the alkyl-nickel bond in methyl-coenzyme M reductase. Biochemistry. 2008;47:2661–2667. doi: 10.1021/bi701942w. [DOI] [PubMed] [Google Scholar]
- 29.Pelmenschikov V, Siegbahn PE. Catalysis by methyl-coenzyme M reductase: a theoretical study for heterodisulfide product formation. J Biol Inorg Chem. 2003;8:653–662. doi: 10.1007/s00775-003-0461-8. [DOI] [PubMed] [Google Scholar]
- 30.Chen SL, Pelmenschikov V, Blomberg MR, Siegbahn PE. Is there a Ni-methyl intermediate in the mechanism of methyl-coenzyme M reductase? J Am Chem Soc. 2009;131:9912–9913. doi: 10.1021/ja904301f. [DOI] [PubMed] [Google Scholar]
- 31.Bobik TA, Wolfe RS. Physiological importance of the heterodisulfide of coenzyme M and 7-mercaptoheptanoylthreonine phosphate in the reduction of carbon dioxide to methane in Methanobacterium. Proc Natl Acad Sci U S A. 1988;85:60–63. doi: 10.1073/pnas.85.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noll KM, Donnelly MI, Wolfe RS. Synthesis of 7-mercaptoheptanoylthreonine phosphate and its activity in the methylcoenzyme M methylreductase system. J Biol Chem. 1987;262:513–515. [PubMed] [Google Scholar]
- 33.Porat I, Sieprawska-Lupa M, Teng Q, Bohanon FJ, White RH, Whitman WB. Biochemical and genetic characterization of an early step in a novel pathway for the biosynthesis of aromatic amino acids and p-aminobenzoic acid in the archaeon Methanococcus maripaludis. Mol Microbiol. 2006;62:1117–1131. doi: 10.1111/j.1365-2958.2006.05426.x. [DOI] [PubMed] [Google Scholar]
- 34.Telser J. Electron-Nuclear Double Resonance (ENDOR) Spectroscopy. In: Scott RA, Lukehart CM, editors. Applications of Physical Methods to Inorganic and Bioinorganic Chemistry. John Wiley & Sons; Chichester, UK: 2007. pp. 99–124. [Google Scholar]
- 35.Mailer C, Taylor CPS. Rapid adiabatic passage EPR of ferricytochrome c. Signal enhancement and determination of the spin-lattice relaxation time. Biochimica et Biophysica Acta. 1973;322:195–203. doi: 10.1016/0005-2795(73)90293-6. [DOI] [PubMed] [Google Scholar]
- 36.Werst MM, Davoust CE, Hoffman BM. Ligand Spin Densities in Blue Copper Proteins by Q-band 1H and 14N ENDOR Spectroscopy. Journal of the American Chemical Society. 1991;113:1533–1538. [Google Scholar]
- 37.Davoust CE, Doan PE, Hoffman BM. Q-Band Pulsed Electron Spin-Echo Spectrometer and Its Application to ENDOR and ESEEM. Journal of Magnetic Resonance. 1996;119:38–44. [Google Scholar]
- 38.Lee H-I, Igarashi RY, Laryukhin M, Doan PE, Dos Santos PC, Dean DR, Seefeldt LC, Hoffman BM. An Organometallic Intermediate during Alkyne Reduction by Nitrogenase. Journal of the American Chemical Society. 2004;126:9563–9569. doi: 10.1021/ja048714n. [DOI] [PubMed] [Google Scholar]
- 39.Doan PE, Gurbiel RJ, Hoffman BM. The Ups and Downs of Feher-Style ENDOR. Appl. Magn. Reson. 2007;31:647–661. [Google Scholar]
- 40.Schweiger A, Jeschke G. Principles of Pulse Electron Paramagnetic Resonance. Oxford University Press; Oxford, UK: 2001. [Google Scholar]
- 41.Krymov V, Gerfen GJ. Analysis of the tuning and operation of reflection resonator EPR spectrometers. J Magn Reson. 2003;162:466–478. doi: 10.1016/s1090-7807(03)00109-5. [DOI] [PubMed] [Google Scholar]
- 42.Ranguelova K, Girotto S, Gerfen GJ, Yu S, Suarez J, Metlitsky L, Magliozzo RS. Radical sites in Mycobacterium tuberculosis KatG identified using electron paramagnetic resonance spectroscopy, the three-dimensional crystal structure, and electron transfer couplings. J Biol Chem. 2007;282:6255–6264. doi: 10.1074/jbc.M607309200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harmer J, Finazzo C, Piskorski R, Bauer C, Jaun B, Duin EC, Goenrich M, Thauer RK, Doorslaer SV, Schweiger A. Spin Density and Coenzyme M Coordination Geometry of the ox1 Form of Methyl-Coenzyme M Reductase: A Pulse EPR Study. J. Am. Chem. Soc. 2005;127:17744–17755. doi: 10.1021/ja053794w. [DOI] [PubMed] [Google Scholar]
- 44.Craft JL, Horng Y-C, Ragsdale SW, Brunold TC. Nickel Oxidation States of F430 Cofactor in Methyl-Coenzyme M Reductase. Journal of the American Chemical Society. 2004;126:4068–4069. doi: 10.1021/ja038082p. [DOI] [PubMed] [Google Scholar]
- 45.Finazzo C, Harmer J, Jaun B, Duin EC, Mahlert F, Thauer RK, Doorslaer SV, Schweiger A. Characterization of the MCRred2 form of methyl-coenzyme M reductase: a pulse EPR and ENDOR study. J. Biol. Inorg. Chem. 2003;8:586–593. doi: 10.1007/s00775-003-0450-y. [DOI] [PubMed] [Google Scholar]
- 46.Hyde JS, Froncisz W. The Role of Microwave Frequency in EPR Spectroscopy of Copper Complexes. Ann. Rev. Biophys. Bioeng. 1982;11:391–417. doi: 10.1146/annurev.bb.11.060182.002135. [DOI] [PubMed] [Google Scholar]
- 47.Harmer J, Finazzo C, Piskorski R, Ebner S, Duin EC, Goenrich M, Thauer RK, Reiher M, Schweiger A, Hinderberger D, Jaun B. A Nickel Hydride Complex in the Active Site of Methyl-Coenzyme M Reductase: Implications for the Catalytic Cycle. J. Am. Chem. Soc. 2008;130:10907–10920. doi: 10.1021/ja710949e. [DOI] [PubMed] [Google Scholar]
- 48.Dorlet P, Valentin MD, Babcock GT, McCracken JL. Interaction of YZ· with Its Environment in Acetate-Treated Photosystem II Membranes and Reaction Center Cores. J. Phys. Chem. B. 1998;102:8239–8247. [Google Scholar]
- 49.Hoganson CW, Babcock GT. Protein-tyrosyl radical interactions in photosystem II studied by electron spin resonance and electron nuclear double resonance spectroscopy: comparison with ribonucleotide reductase and in vitro tyrosine. Biochemistry. 1992;31:11874–11880. doi: 10.1021/bi00162a028. [DOI] [PubMed] [Google Scholar]
- 50.Shi W, Hoganson CW, Espe M, Bender CJ, Babcock GT, Palmer G, Kulmacz RJ, Tsai A.-l. Electron Paramagnetic Resonance and Electron Nuclear Double Resonance Spectroscopic Identification and Characterization of the Tyrosyl Radicals in Prostaglandin H Synthase 1. Biochemistry. 2000;39:4112–4121. doi: 10.1021/bi992561c. [DOI] [PubMed] [Google Scholar]
- 51.Dorlet P, Seibold SA, Babcock GT, Gerfen GJ, Smith WL, Tsai A.-l., Un S. High-Field EPR Study of Tyrosyl Radicals in Prostaglandin H2 Synthase-1. Biochemistry. 2002;41:6107–6114. doi: 10.1021/bi015871f. [DOI] [PubMed] [Google Scholar]
- 52.Hoganson CW, Sahlin M, Sjöberg B-M, Babcock GT. Electron Magnetic Resonance of the Tyrosyl Radical in Ribonucleotide Reductase from Escherichia coli. J. Am. Chem. Soc. 1996;118:4672–4679. [Google Scholar]
- 53.Whittaker JW. Free Radical Catalysis by Galactose Oxidase. Chem. Rev. 2003;103:2347–2364. doi: 10.1021/cr020425z. [DOI] [PubMed] [Google Scholar]
- 54.Gerfen GJ, Bellew BF, Griffin RG, Singel DJ, Ekberg CA, Whittaker JW. High-Frequency Electron Paramagnetic Resonance Spectroscopy of the Apogalactose Oxidase Radical. J. Phys. Chem. 1996;100:16739–16748. [Google Scholar]
- 55.Stubbe J, van Der Donk WA. Protein Radicals in Enzyme Catalysis. Chem Rev. 1998;98:705–762. doi: 10.1021/cr9400875. [DOI] [PubMed] [Google Scholar]
- 56.Wagner AF, Frey M, Neugebauer FA, Schafer W, Knappe J. The free radical in pyruvate formate-lyase is located on glycine-734. Proc Natl Acad Sci U S A. 1992;89:996–1000. doi: 10.1073/pnas.89.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sono M, Roach MP, Coulter ED, Dawson JH. Heme-Containing Oxygenases. Chem. Rev. 1996;96:2841–2888. doi: 10.1021/cr9500500. [DOI] [PubMed] [Google Scholar]
- 58.Frey PA, Hegeman AD, Reed GH. Free radical mechanisms in enzymology. Chem Rev. 2006;106:3302–3316. doi: 10.1021/cr050292s. [DOI] [PubMed] [Google Scholar]
- 59.Symons MCR. On the electron spin resonance detection of RS radicals in irradiated solids: radicals of type RSSR−, RS-SR+ 2, and R2SSR2+ J. Chem. Soc., Perkin Trans. 2. 1974:1618–1620. [Google Scholar]
- 60.Gastel M. v., Lubitz W, Lassmann G, Neese F. Electronic Structure of the Cysteine Thiyl Radical: A DFT and Correlated ab Initio Study. J. Am. Chem. Soc. 2004;126:2237–2246. doi: 10.1021/ja038813l. [DOI] [PubMed] [Google Scholar]
- 61.Lassmann G, Kolberg M, Bleifuss G, Gräslund A, Sjöberg B-M, Lubitz W. Protein thiyl radicals in disordered systems: A comparative EPR study at low temperature. Phys. Chem. Chem. Phys. 2003;5:2442–2453. [Google Scholar]
- 62.Larsson A, Sjoberg BM. Identification of the stable free radical tyrosine residue in ribonucleotide reductase. EMBO J. 1986;5:2037–2040. doi: 10.1002/j.1460-2075.1986.tb04461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petersson L, Graslund A, Ehrenberg A, Sjoberg BM, Reichard P. The iron center in ribonucleotide reductase from Escherichia coli. J Biol Chem. 1980;255:6706–6712. [PubMed] [Google Scholar]
- 64.Svistunenko DA. Reaction of haem containing proteins and enzymes with hydroperoxides: the radical view. Biochim Biophys Acta. 2005;1707:127–155. doi: 10.1016/j.bbabio.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 65.Cheesman MR, Ankel-Fuchs D, Thauer RK, Thomson AJ. The magnetic properties of the nickel cofactor F430 in the enzyme methyl-coenzyme M reductase of Methanobacterium thermoautotrophicum. Biochem. J. 1989;260:613–616. doi: 10.1042/bj2600613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.