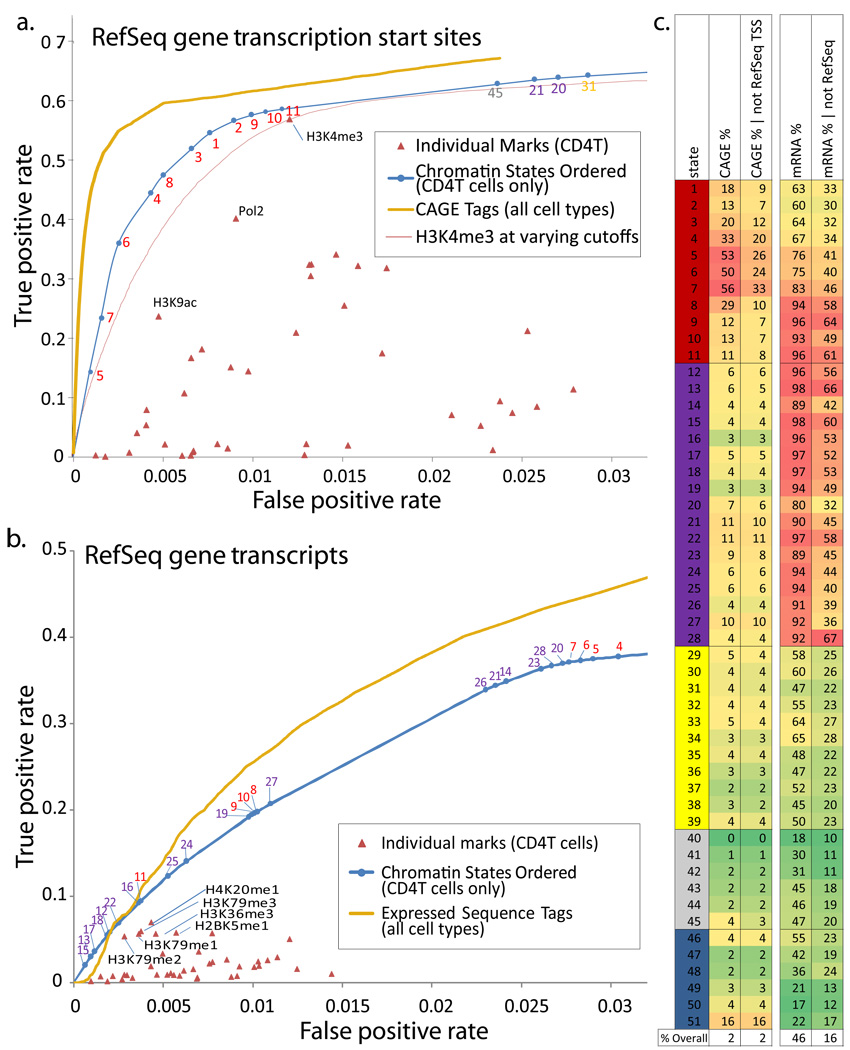
Figure 5. Discovery power of chromatin states for genome annotation.
a. Comparison of the power to discover transcription start sites (TSS) for individual chromatin marks (red), chromatin states (blue) ordered by their TSS enrichment, and a directed experimental approach based on CAGE sequence tag data read counts from all available cell types48 (gold), while the chromatin-states and marks only use data from CD4 T-cells. Both chromatin states and CAGE tags are compared using a Receiver Operating Characteristic (ROC) curve that shows the false positive (x-axis) and true positive (y-axis) rates at varying prediction thresholds in the task of predicting if a 200bp interval intersects a RefSeq TSS. Thin red curve compares performance of H3K4me3 mark at varying intensity thresholds. b. Comparison of the power to detect RefSeq transcribed regions for chromatin states and marks as in a, and directed experimental information coming from Expressed Sequence Tag (EST) data (gold) based on sequence counts from all available cell types38, 49. c. Independent experimental and comparative information provides support that a significant fraction of ‘false positives’ in panels a and b are genuine novel unannotated TSS and transcribed regions currently missing from RefSeq. Percentage of each state supported by a CAGE tag (column 1), and the same percentage for locations at least 2kb away from a RefSeq TSS (column 2), suggests that many promoter-associated states outside RefSeq promoters are supported by CAGE tag evidence. Similarly, percentage of each state overlapping a GenBank mRNA (column 3), and the same percentage specifically outside RefSeq genes (column 4), suggest that transcription-associated states outside RefSeq genes are supported by mRNA evidence. Similar support is found by GenBank Expressed Sequence Tags (ESTs) and evolutionarily-conserved predicted new exons (Supplementary Fig. 33).