Abstract
Rad51 protein promotes homologous recombination in eukaryotes. Recombination activities are activated by Rad51 filament assembly on ssDNA. Previous studies of yeast Rad51 showed that His352 occupies an important position at the filament interface, where it could relay signals between subunits and active sites. To investigate, we characterized yeast Rad51 H352A and H352Y mutants, and solved the structure of H352Y. H352A forms catalytically competent but salt-labile complexes on ssDNA. In contrast, H352Y forms salt-resistant complexes on ssDNA, but is defective in nucleotide exchange, RPA displacement and strand exchange with full-length DNA substrates. The 2.5 Å crystal structure of H352Y reveals a right-handed helical filament in a high-pitch (130 Å) conformation with P61 symmetry. The catalytic core and dimer interface regions of H352Y closely resemble those of DNA-bound Escherichia coli RecA protein. The H352Y mutation stabilizes Phe187 from the adjacent subunit in a position that interferes with the γ-phosphate-binding site of the Walker A motif/P-loop, potentially explaining the limited catalysis observed. Comparison of Rad51 H352Y, RecA–DNA and related structures reveals that the presence of bound DNA correlates with the isomerization of a conserved cis peptide near Walker B to the trans configuration, which appears to prime the catalytic glutamate residue for ATP hydrolysis.
INTRODUCTION
Homologous recombination (HR) is essential for the accurate repair of DNA double-strand breaks and for the maintenance of genomic stability. HR also provides a key mechanism for the generation of genetic diversity. In eukaryotes, the central steps of HR require the ATP-dependent DNA strand exchange activity of Rad51 protein, which is a member of the highly conserved RecA recombinase family (1). DNA strand exchange reactions occur between homologous molecules of single- and double-stranded DNA. The first step of DNA strand exchange is the formation of a competent presynaptic filament of Rad51 protein on single-stranded DNA. Filament assembly allosterically activates the enzymatic activities of Rad51 including ssDNA-dependent ATP hydrolysis, homology search and DNA strand exchange. The proper assembly and activation of Rad51–ssDNA filaments are critical for DSB repair and cancer avoidance, as illustrated by defects in this process that are associated with cancer predisposition syndromes in humans (2).
Presynaptic filament assembly, turnover and DNA strand exchange activity are coupled to the binding and hydrolysis of ATP by Rad51. Therefore signaling between binding sites for DNA and nucleotide ligands is critical for Rad51 recombination activity. It is known that ATP induces or stabilizes a high-affinity ssDNA-binding conformation of Rad51 that is required for the assembly of catalytically competent presynaptic filaments (3). The hydrolysis of ATP to ADP weakens Rad51–DNA interactions, facilitating filament turnover. In the RecA/Rad51 family, high-affinity binding to ssDNA in the presence of ATP or of non-hydrolyzable ATP analogs correlates with a high-pitch filament morphology detected by electron microscopy and crystallographic methods (4,5). The high-pitch filament is generally referred to as the active form of the filament. Low-affinity binding to ssDNA in the presence of ADP or no nucleotide correlates with a low-pitch filament morphology that is generally referred to as the inactive form of the filament. The mechanism of inter-conversion between these forms, if it occurs during recombination, is unknown.
The assembly of Rad51 filaments on ssDNA is modulated by RPA, the eukaryotic ssDNA-binding protein. RPA promotes filament assembly by removing regions of secondary structure from ssDNA that would otherwise impede Rad51 binding, but the high affinity of RPA for ssDNA imposes a potential barrier to Rad51 filament assembly in vitro (6,7). Biochemical data demonstrate that to form a competent presynaptic filament, Rad51 must displace RPA in order to bind to sites on ssDNA. In vitro, strand exchange occurs optimally when Rad51 is pre-incubated with ssDNA and ATP prior to RPA addition. When the ssDNA is pre-saturated with RPA, however, as is likely to occur in vivo, strand exchange is inhibited due to the rate-limiting displacement of RPA from ssDNA by Rad51. The inhibition is reversed by recombination mediator proteins (Rad52, Rad51 paralogs, or Brca2) that promote RPA displacement by Rad51 (2,8). The importance of Rad51/RPA competition in recombination is illustrated by the observation that recombination-deficient mutations in yeast recombination mediator proteins Rad55 and Rad57 are complemented by mutations in Rad51 that increase its ssDNA-binding affinity and therefore its ability to compete with RPA for binding sites on ssDNA (9).
The coupling of ssDNA binding to catalysis is of central importance in the recombination activities of all Rad51/RecA enzymes, but the mechanism of signaling between the various DNA-bound states and catalytic sites is poorly understood. This signaling to and from the active site is coordinated by loops L1 and L2, which are disordered in recombinase structures lacking DNA, but ordered in RecA–DNA structures (4). Major ssDNA binding determinants occur in L2, which is connected at its N-terminal end to a conserved glutamine residue (Gln194 in Escherichia coli RecA, Gln326 in Saccharomyces cerevisiae Rad51) that is proposed to act as a sensor for the presence of ATP in the active site (10). Other interactions that span the protomer–protomer interface are proposed to play key roles in coordinating DNA binding to the activation of catalysis. Crystal structures of high-pitch filaments show that the ATPase active site itself spans the protomer–protomer interface (4,11). In the RecA–DNA structures, Lys248 and Lys250 from one protomer coordinate the γ-phosphate of ATP bound to the P-loop region of the adjacent protomer, with Lys250 interacting with the putative catalytic glutamate residue of the adjacent protomer (4). These interactions are proposed to stabilize the interface and activate hydrolysis in response to DNA binding (4). This mechanism is unlikely to be conserved outside of the eubacteria, however, since the sequence and structural motifs containing the two lysine residues are missing from eukaryotic Rad51 and archaeal RadA enzymes.
To date only one structure of a eukaryotic Rad51 filament has been published. The S. cerevisiae Rad51 I345T mutant is a gain-of-function mutant with enhanced ssDNA-binding affinity (9). The crystallographic structure of I345T revealed a filament with P31 symmetry (11). An interesting feature of the I354T structure is that the repeating unit is a dimer, so that alternating momomers are in different conformations, creating two classes of interfaces and thus two classes of ATPase active sites. The I354T structure shows that residue His352, which is highly conserved among Rad51, Dmc1 and RadA recombinases, occupies an important position at the protomer–protomer interface near the ATPase site. The equivalent residue in E. coli RecA is Phe217, which is known to be important for allosteric communication within RecA–ssDNA filaments (12). In yeast Rad51 filaments, His352 interacts with P-loop/Walker A residue Phe187 from the adjacent protomer. His352–Phe187 interactions occur in two distinct geometries at alternating interfaces within the 31 filament (11). The relative positioning of His352 and Phe187 appears to be coupled to the movements of other elements of the interface and ATPase sites. Therefore His352 is a strong candidate for a residue that couples ssDNA binding and filament formation with the nucleotide state of Rad51 protein. Consistent with this idea, site-directed mutagenesis of His352 reduces the ability of Rad51 to perform DNA strand exchange between simple oligonucleotide substrates (13).
In this study we examined the biochemical defects caused by two different site-directed mutations at the His352 locus of S. cerevisiae Rad51 protein, H352A and H352Y, and we solved the X-ray crystallographic structure of the H352Y protein filament. We find that these mutants are defective in DNA strand exchange but for different reasons: H352A has the hallmarks of a classic weak ssDNA binder that forms catalytically competent but salt-labile complexes on ssDNA. In contrast the H352Y mutant is a strong ssDNA binder, but is unable to undergo multiple turnovers of the nucleotide state. ATP hydrolysis by H352Y appears to be limited to a single turnover, consistent with a defect in nucleotide exchange (replacement of ADP product with new ATP substrate). The results indicate that cross-interface interactions mediated by His352 are important for normal signaling between ssDNA-binding and catalytic sites in Rad51. The filament structure reveals that His → Tyr substitution at position 352 stabilizes Phe187 from the adjacent subunit in a position that interferes with the γ-phosphate-binding site of Walker A, potentially explaining the limited catalysis observed in H352Y. Comparison of the Rad51 H352Y structure to other RecA/Rad51 structures suggests that Phe187 (Glu68 in E. coli RecA) of Walker A either plays a role in detecting the presence/absence of the γ-phosphate of ATP, or in preventing nucleotide exchange. Comparison further reveals that the presence of bound DNA correlates with the isomerization of a conserved cis peptide at the end of the Walker B motif to the trans configuration, which appears to prime the putative nucleophilic activator Glu221 (Glu96 in E. coli RecA) for hydrolysis of the ATP.
MATERIALS AND METHODS
Reagents
Chemicals, biochemicals and enzymes were purchased from Sigma-Aldrich unless specifically stated. All restriction enzymes were purchased from New England Biolabs. The crystallization screens, trays and plates were purchased from Hampton Research. All reagents were analytical grade and solutions were made with Barnstead NANO-pure water. TE buffer contained 10 mM Tris–HCl, pH 8.0 and 1 mM EDTA.
Nucleic acids
Circular single-stranded φX174 virion DNA (5.4 kb) and double-stranded φX174 DNA (replicative form I DNA, RFI) were purchased from New England Biolabs. RFI was linearized by digestion with PstI restriction endonuclease and purified using standard procedures. The concentrations of ssDNA and dsDNA stock solutions were determined according to manufacturer’s instructions. Etheno-modified ssDNA (εDNA) was prepared from M13mp19 virion ssDNA (7.3 kb) and quantified as previously described (14,15). All DNA molecules were stored at −20°C in TE buffer and the concentrations were expressed in units of micromoles of nucleotide residues per liter.
Site-directed mutagenesis of yeast Rad51 protein
Plasmid pLant2B (RIL) encoding yeast Rad51 was a generous gift from Dr Tom Ellenberger at Washington University in St Louis. Two mutants, H352A and H352Y at residue 352 of Rad51 protein were introduced using the QuikChange site-directed mutagenesis protocol (Stratagene). To construct the Rad51–H352A mutant, forward primer 1, 5′GTAATATTATGGCAGCGTCTTCCACCACGCGAT3′ and reverse primer 2, 5′ATCGCGTGGTGGAAGACGCTGCCATAATATTAC3′ were designed. Similarly, forward primer 3, 5′GTGGTAATATTATGGCATATTCTTCCACCACGCG3′ and reverse primer 4, 5′CGCGTGGTGGAAGAATATGCCATAATATTACCAC3′ were used for introducing the Rad51–H352Y mutant. PCR reactions were conducted according to the QuikChange protocol and the resulting plasmids were sequenced at the Vermont Cancer Center DNA Sequencing Facility to verify successful mutagenesis.
Purification of yeast RPA and Rad51 proteins
Yeast RPA protein expression plasmid p11d-tRPA was a kind gift from Dr Mark Wold of the University of Iowa. RPA was expressed in E. coli strain BL21(DE3) and the protein was purified as described (16).
Rad51 proteins were expressed from plasmid pLant2B (RIL) harboring wild-type or mutant versions of the RAD51 gene, which was transformed into E. coli strain BLR(DE3)/pLysS (Novagen, Inc., Madison, WI). Identical purification protocols were used for Rad51 wild-type, H352A and H352Y proteins. The purification steps were a modified version of a published protocol (17). In brief, the cells were grown at 37°C to a cell density of A600 = 0.6. Cells were shifted to 16°C and induced with 0.1 mM IPTG for 16 h. Cell pellets from 6-l cultures were harvested by centrifugation and resuspended in 120 ml of Buffer A containing 600 mM KCl, 50 mM Tris–HCl (pH 7.5), 5 mM EDTA, 10% sucrose, 10 mM β-mercaptoethanol, 0.1 mM benzamidine and 5 µg/ml 4-(2-aminoethyl) benzenesulfonyl fluoride (AEBSF, Sigma). Cells were lysed by sonication, and extracts were clarified by centrifugation (40 000 rpm, 2 h). The extract was treated with ammonium sulfate (0.22 g/ml), and the protein pellet was resuspended in 120 ml of Buffer P [20 mM potassium phosphate (pH 7.5), 10% glycerol, 0.5 mM EDTA, 0.5 mM DTT], containing 50 mM KCl, 5 μg/ml AEBSF and 0.1 mM benzamidine. This solution was loaded at a flow rate of 120 ml/h onto a 50-ml Cybacron Blue-agarose column pre-equilibrated with Buffer P. The column was washed with Buffer P containing 100 mM KCl until the A280 of the flow-through approached zero, and then 200 ml of a 0.2–1.0 M KCl gradient in Buffer P was applied. Fractions containing Rad51 protein were combined and dialyzed against Buffer P containing 100 mM KCl, 5 μg/ml AEBSF and 0.1 mM benzamidine, and then loaded onto a 50-ml Q-Sepharose column, which was pre-equilibrated with Buffer P containing 100 mM KCl. Proteins were fractionated with a 600 ml KCl gradient (200–800 mM KCl) in Buffer P; Rad51 protein eluted at about 400 mM KCl. Pooled fractions were dialyzed against Buffer H [5 mM potassium phosphate (pH 6.5), 0.5 mM DTT, 10% glycerol, 40 mM KCl] containing 5 μg/ml AEBSF and 0.1 mM benzamidine, and then loaded onto a 50-ml Bio-Gel hydroxyapatite column that was equilibrated with Buffer H. Bound proteins were eluted with a 300 ml linear gradient of 20–200 mM potassium phosphate (pH 6.5) in Buffer H at a flow rate of 60 ml/h. These fractions were pooled and dialyzed against Buffer P containing 200 mM KCl and then loaded onto a MonoQ (HR 5/5) column. A linear gradient (160 ml) of 200–600 mM KCl in Buffer P was used to elute Rad51 protein (at about 400 mM KCl). Fractions containing purified Rad51 protein were dialyzed against storage buffer [20 mM Tris–HCl (pH 7.5), 1 mM DTT, 10% glycerol, 0.5 mM EDTA, 100 mM KCl], aliquoted, and stored at −80°C. The final yield of wild type Rad51 protein was about 25 mg from 6 l of culture. The concentrations of WT and H352A proteins were calculated using an extinction coefficient (determined from amino-acid composition) of 1.29 × 104 M–1cm–1 at 280 nm. H352Y protein concentration was calculated using an extinction coefficient (determined from amino-acid composition) of 1.4 × 104 M–1cm–1 at 280 nm. The purity of Rad51 protein was above 99% based on SDS–polyacrylamide gel electrophoresis. The presence of tyrosine at position 352 of the H352Y mutant was confirmed by mass spectrometry at the Proteomics Facility of the Vermont Genetics Network, University of Vermont College of Medicine.
All protein stock solutions were tested for contaminating nuclease activities by the following procedures (18): Aliquots of each protein stock were incubated for 2 h at 37°C in solutions containing either circular ssDNA, linear ssDNA, linear dsDNA, or supercoiled plus nicked plasmid dsDNA, in buffer used for DNA strand exchange assays (see below). Samples were then deproteinized, electrophoresed on agarose gels, and visualized by ethidium bromide staining as described below for DNA strand exchange assays. The gels were examined visually for evidence of nuclease degradation of the various DNA species (smearing, appearance of low molecular weight species, nicking or linearization of circular DNA species). All protein stocks used in this study were found to be nuclease-free according to these criteria.
Etheno-DNA (εDNA) fluorescence enhancement assays for Rad51–ssDNA interactions
The binding site sizes of Rad51 wild-type, H352A and H352Y proteins on single-stranded DNA were determined from titrations of εDNA with protein under low-salt, tight binding conditions, while monitoring the increase in εDNA fluorescence as previously described (17). Fluorescence measurements were made in a PTI Quantamaster QM-6 fluorometer (Photon Technology International, South Brunswick, NJ) at excitation and emission wavelengths of 300 nm and 405 nm, respectively. The bandwidths for excitation and emission were set at 2 nm and 5 nm, respectively. Titrations were carried out at 37°C in a quartz cuvette containing a starting volume of 700 µl of solution consisting of 3 µM εDNA, 2 mM ATP, 30 mM Tris–acetate (pH 7.5), 10 mM magnesium acetate and 0.1 mM DTT. Control titrations of protein into buffer lacking εDNA were used to correct for the contribution of intrinsic tryptophan fluorescence to signal. All data were corrected for dilution. Photobleaching and inner filter effects were assessed using standard procedures (19) and found to be negligible.
Electrophoretic mobility shift assays for Rad51–ssDNA interactions
Rad51–ssDNA interactions were analyzed by native agarose gel electrophoretic mobility shift assay (EMSA). A constant concentration of 30 µM φX174 ssDNA was mixed with varying amounts of wild-type or mutant Rad51 protein in 30 mM Tris–acetate (pH 7.5), 5 mM MgCl2, 0 or 2 mM ATP depending on the experiment and 0–1000 mM NaCl depending on the experiment, in a total volume of 20 µl and incubated at 37°C for 30 min. Immediately after incubation sample loading buffer [3 µl of 40 mM Tris–acetate (pH 7.5), 50% glycerol and a trace of bromphenol blue] was added to each reaction, and the protein–ssDNA complexes were separated from unbound ssDNA by electrophoresis through 0.8% agarose gels in TAE buffer (40 mM Tris–acetate, pH 8.0, 2 mM EDTA) at 120 V for 2 h and were visualized by ethidium bromide staining.
EMSAs for nucleotide binding
The binding of radiolabeled nucleotide (α-[32P]-ATP or -ADP) to Rad51 in the presence/absence of ssDNA was monitored by native polyacrylamide gel EMSA. In total reaction volumes of 20 µl, a constant concentration of 10 µM α-[32P]-ATP (10 µCi/ml final specific activity) was incubated with 10 µM wild-type or mutant Rad51 protein in the presence or absence of 30 µM φX174 ssDNA. Reaction buffer contained 30 mM Tris–acetate (pH 7.5), 10 µg/ml BSA, 1 mM DTT, 10 mM magnesium acetate, 30 mM KCl and 5% glycerol. Otherwise identical control reactions lacking protein, lacking both ssDNA and protein, or lacking nucleotide (background control) were carried out in parallel. Reactions mixtures were incubated at 37°C for 2 h. Samples were then loaded onto 12% non-denaturing polyacrylamide gels in a modified buffer containing 30 mM Tris–acetate (pH 7.5), 10 mM magnesium acetate and 30 mM KCl. The gels were made from a 75:1 mixture of acrylamide to bisacrylamide in the modified buffer. Complexes were separated from free nucleotide by electrophoresis at 150 V for 2 h. The gels were dried and exposed to a K-screen (Kodak) and analyzed in a Bio-Rad Personal Molecular Imager-FX with Quantity One software v4.5.1. Concentrations of bound and free nucleotide (Nbound and Nfree, respectively) were calculated according to the equations Nbound = NtotalIbound/(Ibound + Ifree) and Nfree = NtotalIfree/(Ibound + Ifree), where Ntotal is the total nucleotide concentration (10 µM), Ibound is the phosphorimage intensity of the bound nucleotide spot in a given lane, and Ifree is the phosphorimage intensity of the free nucleotide spot in the same lane.
ATPase assays
Steady-state rates of ATP hydrolysis were measured using thin-layer chromatography (TLC) assays. Reactions at 37°C contained 30 mM MES (pH 6.2), 6 mM MgCl2, 30 mM KCl, 1 mM DTT and 0.1 mg/ml BSA in a final volume of 50 µl. The monovalent salt concentration was increased to 80 mM in certain reactions by the addition of 50 mM NaCl. In addition the reaction mixtures contained 2.0 µM wild-type or mutant Rad51 protein and 12 µM φX174 ssDNA. Reactions were initiated by the addition of α-[32P]-ATP to a final concentration of 3 mM (10 µCi/ml final specific activity). Aliquots (8 µl) were removed at various time points and quenched with 8 mM EDTA and 1% SDS (final concentrations). Quenched samples were spotted out at 1 µl volume onto PEI-cellulose TLC plates (20 × 20 cm) at 1-cm intervals. After all the samples were spotted and dried the TLC plates were developed with 0.75 M KH2PO4 and allowed to air dry. The TLC plates were exposed for 1.5 h to a K-screen (Kodak) and scanned by a Bio-Rad Personal Molecular Imager-FX. Quantification of the phosphorimage was performed by Quantity One v4.5.1 (Bio-Rad) software and subsequently fit using KaleidaGraph v 3.6.2 (Synergy Software). The TLC assay was also used to measure the hydrolysis of ATP in samples identical to those used in nucleotide-binding EMSA experiments. Following incubation at 37°C for 2 h, 8 µl aliquots were removed and quenched with 8 mM EDTA and 1% SDS, spotted onto TLC plates and developed as described above.
DNA strand exchange assays
DNA strand exchange reactions using φX174 DNA substrates were carried out as described (20) with minor changes. Reactions were assembled as follows (final volume 12.5 µl, final concentrations are given): 11 µM wild-type or mutant Rad51 protein, 33 µM circular φX174 ssDNA and 2.5 mM ATP were preincubated for 5 min at 37°C in buffer containing 35 mM MOPS (pH 7.2), 100 µg/ml bovine serum albumin, 1 mM DTT, 3 mM MgCl2 and 10 mM KCl. To each reaction 1.65 µM RPA was added and the mixture was incubated at 37°C for another 10 min. Finally, 33 µM PstI-linearized φX174 dsDNA and 4 mM spermidine (pH 7.2) were added to start the reaction. After incubation at 37°C for the indicated time, the reactions were stopped by adding SDS and proteinase K to final concentrations of 1% and 0.5 mg/ml, respectively, followed by 15-min incubation at 37°C. The deproteinized samples were mixed with loading buffer (3 µl of 40 mM Tris–acetate, pH 7.5, 50% glycerol and a trace of bromphenol blue) and electrophoresed on a 0.8% agarose gel in TAE buffer at 120 V for 2 h. The DNA substrates and strand exchange products were visualized by exposure to ultraviolet light after staining by ethidium bromide. Results were quantified by measuring the intensity of the nicked circle product band and joint molecules in a Bio-Rad Personal Molecular Imager-FX using Quantity One v4.5.1 software.
RPA displacement assays
The ability of wild-type and mutant Rad51 proteins to displace RPA from ssDNA was monitored by the recovery of intrinsic RPA tryptophan fluorescence in the free compared with the ssDNA-bound state as described (21). RPA fluorescence was monitored continuously in the PTI Quantamaster QM-6 fluorometer at excitation and emission wavelengths of 290 and 345 nm, respectively. The bandwidths for excitation and emission were set at 2 and 5 nm, respectively. Reactions were carried out at 37°C in a quartz cuvette containing 700 µl final volume of 30 mM Tris–acetate (pH 7.5), 5 mM magnesium acetate, 50 mM KCl, 1 mM DTT, 2.5 mM ATP, 0.25 µM RPA and 5 µM φX174 ssDNA. The monovalent salt concentration was increased to 100 mM in certain experiments by the addition of 50 mM NaCl. This mixture was pre-incubated for 3 min while monitoring the starting fluorescence signal of RPA–ssDNA. Then the reaction was started by adding 2 µM of wild-type or mutant Rad51 and resulting changes in fluorescence signal were monitored over time. Data were corrected for the residual fluorescence of Rad51 protein and for the effects of dilution, photobleaching and inner filter effects as described (18,22,23). A control reaction lacking Rad51 but containing all other components including Rad51 storage buffer was used to establish the minimum fluorescence signal of the RPA–ssDNA mixture (F0), which was subtracted from all data. A separate control reaction lacking ssDNA was used to establish the maximum fluorescence signal of free RPA, against which all data were normalized. Thus the value F – F0 has a minimum value of zero representing RPA that is fully saturated with ssDNA, and a maximum value of one representing RPA that is completely displaced from ssDNA.
Crystallization procedures
Rad51 H352Y crystals were grown in hanging drop by vapor diffusion. Rad51 H352Y protein from storage buffer was dialyzed into 20 mM Tris–HCl (pH 7.5), 1 mM DTT and concentrated to 20 mg/ml using vivaspin (Sartorius Stedim Biotech) ultrafiltration spin columns. By mixing 1:1 (v/v) of well solution and protein solution the protein was crystallized using the hanging drop method at a temperature of 18°C. Optimal yet small crystals were obtained using reservoir solution containing 35% (v/v) pentaerythritol propoxylate (5/4 PO/OH), 300 mM KCl and 50 mM HEPES (pH 7.5 adjusted by KOH).
X-ray diffraction and structure determination
Diffraction data were collected at beamline 23-ID-B at the APS synchrotron at a wavelength of 1.03 Å, and reduced using HKL2000 (HKL Research) (24) with statistics given in Table 1. EPMR 8.10 (25) readily identified a molecular replacement solution using a protomer of the Rad51–I345T structure (11) (PDB code 1SZP) as the search model. The model, containing a single protomer in the asymmetric unit, was rebuilt with Coot (26) into a composite omit map generated after overall B-factor and rigid-body refinement but prior to any other refinement of the model. After several rounds of crystallographic refinement with CNS 1.21 (27), an alternate conformation of the Tyr352 sidechain was identified and built in. The occupancy of the carboxyl group of Glu221 was reduced to 0.5 based on inspection of residual Fourier maps, although no other single conformation for this group was apparent. The peptide linkage between residues 280–281 was initially modeled as trans; Residual Fourier maps clearly revealed that the linkage was cis, and the model was corrected. A simple modification to the protein parameters file in the standard distribution of CNS 1.21 was necessary to allow for the existence of non-prolyl cis peptide linkages. A peak of strong density seen in the P-loop of the protein was modeled as a chloride ion as there was no phosphate or sulfate in the crystallization mixture. There was no clear density for the N-terminal extension (residues 1–77), nor for the DNA-binding loops 1 (288–293) and 2 (328–347). The model, after inclusion of 79 waters and three chloride ions, gave final refinement statistics as listed in Table 1. A representative Fo – Fc simulated annealing omit map in the region of Tyr352 is shown in Supplementary Figure S1.
Table 1.
Data collection and model refinement statistics
| Data collection | |
| Wavelength (Å) | 1.03320 |
| Space group | P61 |
| Unit cell [a, b, c (Å)] | 78.905, 78.905, 130.333 |
| Resolution (Å) | 50–2.50 (2.59–2.50)a |
| No. of total reflections | 152140 |
| No. of unique reflections | 15937 (1566) |
| Redundancy | 9.6 (5.3) |
| Completeness (%) | 99.7 (98.2) |
| <I>/<σ(I)> | 19.8 (2.0) |
| Rmergeb | 13.0 (66.2) |
| Refinement | |
| No. protein atoms | 2269 |
| No. chloride atoms | 3 |
| No. solvent atoms | 79 |
| Rcrystc | 20.03 |
| Rfreed | 24.57 |
| Estimated coordinate error (Å)e | 0.310/0.385 |
| Deviations from ideal stereochemistry | |
| RMSD bonds (Å) | 0.0063 |
| RMSD angles (°) | 1.347 |
| RMSD B-factors (Å2)f | 1.370/1.825 |
| Average B-factors (Å2)g | 39.4/42.2/56.7 |
| Ramachandran plot analysish | |
| Most favored (%) | 92.0 |
| Additionallly allowed (%) | 7.6 |
| Generously allowed (%) | 0.4 |
| Disallowed (%) | 0.0 |
| PDB ID | 3LDA |
aValues in parentheses are for the highest resolution shell (2.59–2.50 Å).
bRmerge = Σ(|Ii – <I>|)/Σ(I), where Σ is over all reflections measured more than once, and <I> is the mean intensity of all measured observations equivalent to reflection Ii.
cRcryst = Σ(|Fobs – Fcalc|)/Σ(Fobs), where Σ is over all reflections used in refinement, Fobs are the observed diffraction amplitudes, and Fcalc are their corresponding calculated amplitudes from inverse Fourier transformation of the model.
dRfree is defined identically to Rcryst, but involves 9% of the measured reflections not used in refinement and set aside for cross-validation purposes (38).
eEstimated coordinate error is via a Luzzati plot (39) using all reflections and based on cross-validated reflections only.
fRMSD in B-factor between covalently-bonded main chain atoms and side chain atoms.
gAverage B-factors for protein/solvent/chloride ions (40).
hRamacchandran plot analysis is from PROCHECK (41).
RESULTS
Effects of mutations of His352 on Rad51 DNA strand exchange activity
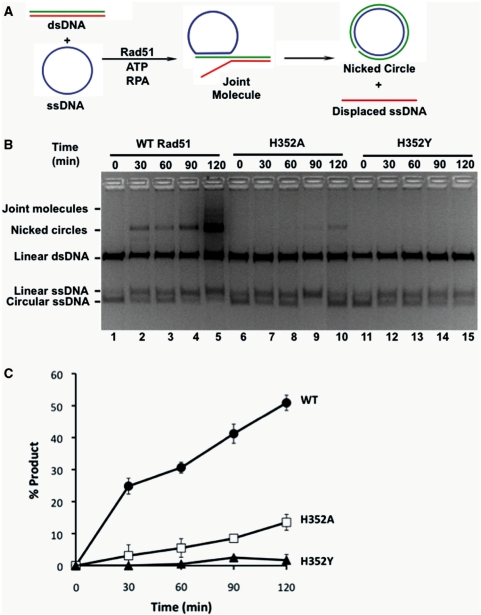
Purified Rad51 H352A, H352Y and wild-type proteins were tested for the ability to catalyze DNA strand exchange reactions using full-length φX174 DNA substrates in the presence of RPA protein. Under these conditions, strand exchange requires the formation of catalytically competent presynaptic filaments on the long (5.4 kb) φX174 ssDNA substrate, which in turn requires Rad51 to successfully compete with RPA for binding sites on ssDNA. A schematic of the DNA strand exchange reaction is shown in Figure 1A. Wild-type Rad51 promotes a strong DNA strand exchange reaction as shown by the appearance of nicked circle and joint molecule products over the 2-h timecourse of the reaction (Figure 1B). The H352A mutant retains DNA strand exchange activity under these conditions, but its activity is much lower than wild-type (Figure 1B). No nicked circle product is visible in reactions with the H352Y mutant (Figure 1B). Strand exchange results were quantified by fluorescence imaging of the nicked circle band (Figure 1C). Results show that the rate of nicked circle product formation catalyzed by H352A is reduced ∼4-fold compared with wild-type, while that of H352Y is reduced by at least 25-fold compared with wild-type.
Figure 1.
DNA strand exchange activities of wild-type and mutant Rad51 proteins. DNA strand exchange assays were carried out using full-length φX174 DNA substrates as described under ‘Materials and Methods’ section. (A) Schematic of in vitro DNA strand exchange reaction, showing interconversion of single- and double-stranded DNA species. (B) Timecourses of DNA strand exchange reactions visualized by separating DNA substrates and products via agarose gel electrophoresis and staining with ethidium bromide. All reactions contained 33 µM each of φX174 circular ssDNA and φX174 linear dsDNA, 11 µM Rad51 enzyme, 1.65 µM RPA and 2.5 mM ATP (final concentrations). Other reaction components and assay conditions are described under ‘Materials and Methods’ section. Lanes 1–5, reaction with wild-type Rad51. Lanes 2–10, reaction with H352A Rad51 mutant. Lanes 11–15, reaction with H352Y Rad51 mutant. (C) Results of panel B quantified by fluorescence imaging. Data are averages from three separate experiments. Filled circles, wild-type Rad51. Open squares, H352A Rad51 mutant. Filled triangles, H352Y Rad51 mutant.
Linear ssDNA formation is a poor indicator of Rad51-catalyzed DNA strand exchange activity, since previous studies showed that the concentration of this species in reaction mixtures is non-stoichiometric with respect to the concentration of nicked circular heteroduplex (34,42). In our experiments, linear ssDNA increases anomalously in reactions with H352A and H352Y (Figure 1B, lanes 6–10 and 11–15), which does not correlate with the observed low rates of heteroduplex formation. We considered that the linear ssDNA present in these reaction mixtures could be the result of a contaminating nuclease activity, although this does not appear to be the case since all of the Rad51 and RPA stocks used in this study were nuclease-free according to sensitive criteria described in ‘Materials and Methods’ section. We conclude that the anomalous appearance of linear ssDNA in these reactions is most likely caused by random strand breaks in the ssDNA that can occur during sample processing steps.
Effects of His352 mutations on Rad51–ssDNA interactions
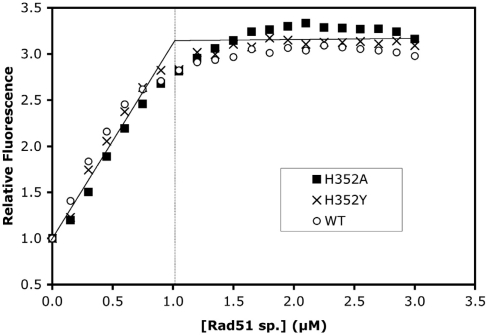
Both H352A and H352Y mutants show severe defects in DNA strand exchange reactions, but the underlying mechanisms could differ in detail. To determine whether changes in protein–ssDNA interactions contribute to strand exchange deficiency, we compared the ssDNA-binding properties of both mutants and wild-type Rad51. The binding site sizes of wild-type, H352A and H352Y on ssDNA were quantified using etheno-DNA (εDNA) fluorescence enhancement assays. Under low-salt conditions, all three enzyme species bind tightly to εDNA in the presence of ATP, and enhance εDNA fluorescence by 3.1- to 3.3-fold at saturation (Figure 2). This degree of fluorescence enhancement is characteristic of proteins that extend and unstack the ssDNA (15,28). All three proteins yielded very similar titration profiles (Figure 2), which were reproducible between different batches of εDNA and protein. Titration endpoints were estimated from the intersection of asymptotic lines drawn through the ascending and plateau portions of each curve, and used to calculate the value of the binding site size parameter, n. The calculated values in nucleotide residues/protomer were n = 3.3 ± 0.3 for wild-type, n = 2.9 ± 0.2 for H352A, and n = 3.1 ± 0.1 for H352Y. Therefore mutations of His352 do not significantly affect the apparent ssDNA-binding site size. The overall average for all three proteins was n = 3.0 ± 0.4, which is indicated by the asymptotic lines drawn in Figure 2, with the vertical line indicating the enzyme concentration at the average endpoint. The measured binding site size of 3-nt residues is similar to previous estimates for wild-type Rad51, including n ≈ 4 from etheno-DNA titrations (17) and n ≈ 3 from DNA strand exchange concentration optima (29).
Figure 2.
ssDNA-binding site size determination for wild-type and mutant Rad51 proteins. Etheno-modified M13mp19 ssDNA (εDNA) was titrated with Rad51 wild-type (open circles), H352A (filled squares), or H352Y (crosses) under low-salt conditions while monitoring the enhancement of εDNA fluorescence as described under ‘Materials and Methods’ section. Solutions contained 3 µM εDNA and 2 mM ATP in buffer consisting of 30 mM Tris–acetate (pH 7.5), 10 mM magnesium acetate and 0.1 mM DTT. The apparent binding site sizes of proteins on this single-stranded lattice are estimated from the titration endpoints. The average endpoint is indicated in this figure by the intersection of the solid asymptotic lines, with the vertical line showing the protein concentration at the average endpoint, corresponding to one Rad51 protomer per three εDNA nucleotide residues.
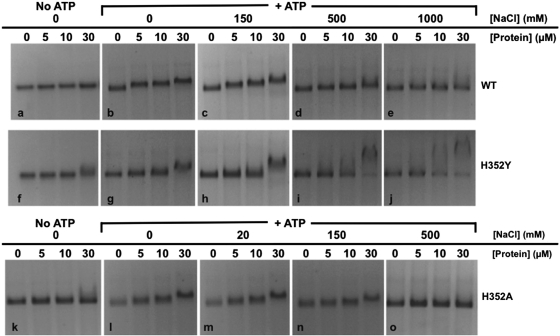
The similar binding site sizes do not rule out the possibility that His352 mutations alter other features of Rad51–ssDNA interactions such as affinity, cooperativity, ATP dependence, or salt effects. Therefore protein interactions with φX174 ssDNA were qualitatively assessed by agarose gel EMSA while varying ATP, salt concentration and protein concentration parameters (Figure 3). The salt-sensitivities of protein–ssDNA interactions provide a means for comparing the relative stabilities of the mutant vs. wild-type complexes. The top row of panels (Figure 3a–e) shows experiments performed with wild-type Rad51. In low-salt buffer containing 30 mM Tris–acetate (pH 7.5), 5 mM MgCl2, and no NaCl, Rad51 complex formation on φX174 ssDNA strongly requires ATP, since little ssDNA mobility shift is observed in its absence (apo conditions—Figure 3a), even at the highest protein concentration where Rad51 is in 3-fold molar excess over potential ssDNA-binding sites. Under otherwise identical conditions, the addition of 2 mM ATP causes a gradual shift to slower migrating species with increasing Rad51 concentration (Figure 3b), indicative of Rad51–ssDNA presynaptic filament formation. These results are consistent with previous reports of ATP-stabilized ssDNA binding by Rad51 (17). In the presence of 2 mM ATP, Rad51–ssDNA complexes are stable at the moderate salt concentration of 150 mM NaCl (Figure 3c), but mobility shift decreases in 500 mM NaCl and is undetectable in 1000 mM NaCl (Figure 3d–e). Therefore wild-type Rad51 forms ATP-stabilized complexes on ssDNA that are moderately salt-resistant.
Figure 3.
Electrophoretic mobility shift assays for ssDNA-binding activities of wild-type and mutant Rad51 proteins—effects of ATP and salt. Protein–ssDNA complexes were assembled at a constant concentration of 30 µM φX174 ssDNA and at variable concentrations of 0–30 µM wild-type or mutant Rad51 enzyme as described under ‘Materials and Methods’ section. Complexes were assembled in buffer containing 30 mM Tris–acetate, pH 7.5, 5 mM MgCl2, with or without ATP and NaCl as indicated. Samples were electrophoresed on agarose gels and visualized by staining with ethidium bromide. Top row—experiments with wild-type Rad51. Nucleotide and salt conditions were as follows: (a) 0 mM ATP (apo conditions), 0 mM NaCl; (b) 2 mM ATP, 0 mM NaCl; (c) 2 mM ATP, 150 mM NaCl; (d) 2 mM ATP, 500 mM NaCl; (e) 2 mM ATP, 1000 mM NaCl. Middle row—experiments with Rad51 H352Y mutant. Nucleotide and salt conditions were as follows: (f) 0 mM ATP (apo conditions), 0 mM NaCl; (g) 2 mM ATP, 0 mM NaCl; (h) 2 mM ATP, 150 mM NaCl; (i) 2 mM ATP, 500 mM NaCl; (j) 2 mM ATP, 1000 mM NaCl. Bottom row—experiments with Rad51 H352A mutant. Nucleotide and salt conditions were as follows: (k) 0 mM ATP (apo conditions), 0 mM NaCl; (l) 2 mM ATP, 0 mM NaCl; (m) 2 mM ATP, 20 mM NaCl; (n) 2 mM ATP, 150 mM NaCl; (o) 2 mM ATP, 500 mM NaCl. Note the lower NaCl concentrations used with H352A compared with H352Y and wild-type Rad51.
The middle row of panels (Figure 3f–j) shows experiments with the Rad51 H352Y mutant. The results demonstrate major differences in the ssDNA-binding properties of H352Y and wild-type. Like wild-type, H352Y forms ATP-stabilized complexes on ssDNA. The ATP requirement is not absolute, however, since a small amount of mobility shift is detectable at the highest H352Y concentration under apo, low-salt conditions (Figure 3f). The addition of 2 mM ATP leads to a much greater degree of ssDNA mobility shift that, remarkably, persists at all salt concentrations tested (0–1000 mM) (Figure 3g–j). This mobility shift is not a result of non-specific aggregation since no material appears in the wells and solutions are non-turbid at all salt concentrations (data not shown). Therefore the H352Y mutation induces a more salt-resistant, higher-affinity form of ssDNA binding than is seen with wild-type Rad51. The H352Y–ssDNA complexes observed at the highest protein concentrations have a tendency to migrate slower with increasing salt concentration (Figure 3h–j), which could be an indication of complex morphology change such as increasing filament stiffness (30,31). H352Y–ssDNA interactions show characteristics of all-or-none binding—this is particularly noticeable at higher protein concentrations in experiments at 500 and 1000 mM NaCl, where highly shifted and free ssDNA co-exist (Figure 3i–j), which raises the possibility of enhanced cooperativity in this mutant. This pattern is never observed with wild-type Rad51, which within its stable salt range shows gradually slower migration with increasing protein concentration, and no free ssDNA, suggesting relatively distributive binding (Figure 3b–d).
The bottom row of panels (Figure 3k–o) shows experiments with the Rad51 H352A mutant. Like wild-type and H352Y, H352A forms ATP-stabilized complexes on ssDNA. Like H352Y, the ATP requirement for H352A–ssDNA binding is not absolute, since a small amount of mobility shift is detectable at the highest H352A concentration under apo, low-salt conditions (Figure 3k). The addition of 2 mM ATP leads to a much greater degree of ssDNA mobility shift, but complex formation occurs only at relatively low salt concentrations, 0–20 mM NaCl (Figure 3l and m). Complex formation is reduced in 150 mM NaCl, and abolished in 500 mM NaCl (Figure 3n and o). Therefore in contrast to H352Y, the H352A mutant exhibits much weaker ssDNA-binding activity than wild-type Rad51. Also, at permissive salt concentrations the binding of H352A resembles the distributive pattern of wild-type more than the apparent cooperative pattern of H352Y.
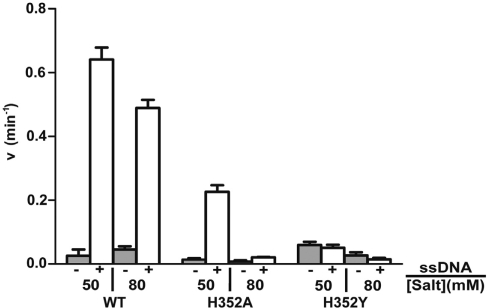
Effects of His352 mutations on steady-state rates of ATP hydrolysis
To further assess the functional defect of H352A and H352Y mutants, steady-state rates of ATPase activity were measured in the presence and absence of ssDNA. Experiments were performed under approximate Vmax conditions for wild-type Rad51—3 mM ATP and a 2-fold excess of φX174 ssDNA-binding sites over enzyme. Reactions were carried out at pH 6.2 to maximize the ATPase activities of H352A and H352Y (13,32). Results are shown in Figure 4. Generally speaking, the relative order of ATPase activity is wild-type > H352A > H352Y, although each activity is condition-dependent as shown in Figure 4. In the presence of 50 mM salt, wild-type Rad51 shows the highest level of ssDNA-dependent ATPase activity, which is 23-fold higher than its basal ATPase rate attained in the absence of ssDNA (Figure 4). The rate of ∼0.65 min–1 measured for wild-type Rad51 under these conditions is consistent with published results (13,33,34). Wild-type Rad51’s ssDNA-dependent ATPase rate decreases only slightly as the salt concentration is increased to 80 mM (Figure 4). Like wild-type, the ATPase activity of H352A is strongly stimulated by ssDNA in 50 mM salt, although its basal and ssDNA-dependent rates are only ∼40% of wild-type (Figure 4). Strikingly, increasing the salt concentration from 50 to 80 mM inhibits H352A’s ssDNA-dependent ATPase activity by ∼12-fold (Figure 4). Therefore the ssDNA-dependent ATPase activity of H352A is dramatically more salt-sensitive than that of wild-type Rad51. The ATPase properties of H352Y differ dramatically from both wild-type and H352A in several ways: The basal rate of hydrolysis in 50 mM NaCl is ∼2.6-fold higher than wild-type, and is not stimulated by ssDNA (Figure 4). In fact ssDNA appears to slightly inhibit ATP hydrolysis by H352Y, as does increasing salt (Figure 4).
Figure 4.
Steady-state rates of ATP hydrolysis by wild-type and mutant Rad51 proteins. ATP hydrolysis was measured by TLC radiometric assay as described under ‘Materials and Methods’ section. Reaction mixtures contained 3 mM ATP and 2.0 µM of either Rad51 wild-type, H352A, or H352Y as indicated below the figure. Reactions were performed either in the absence (gray bars) or in the presence (white bars) of 12 µM φX174 ssDNA. Monovalent salt was added to reactions at the concentrations indicated below the figure. All other buffer components and assay conditions were as described under ‘Materials and Methods’ section.
We note that the ATPase rates observed for Rad51 wild-type, H352A and H352Y in the presence of ssDNA are essentially independent of ssDNA concentration as long as the ssDNA is saturating with respect to enzyme concentration (J. Chen and S. Morrical, unpublished results). We also note that our results differ from those of a previous study wherein a 63-mer oligonucleotide appeared to stimulate the ATPase activity of H352Y (13). To explore this difference, we repeated the ATPase assays in Figure 4 using a 62-mer random-sequence ssDNA oligo in place of 5400-base φX174 ssDNA. The results were essentially identical to those shown in Figure 4 (data not shown). In addition our results were highly reproducible using two different preparations of H352Y. Therefore the different results obtained in our study and (13) are not due to differences in ssDNA length, but could conceivably be due to differences in other reaction parameters such as the higher ATP and magnesium concentrations used in (13).
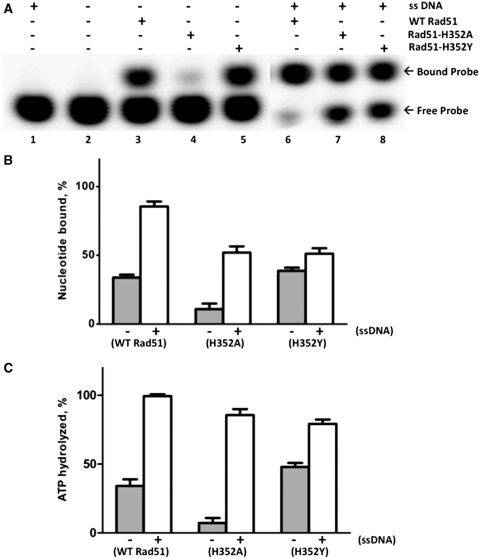
Nucleotide binding and turnover by Rad51 and mutants
We investigated whether the reduced ATPase activities of Rad51 mutants are caused by defects in nucleotide binding or turnover. Experiments were performed under ‘single-turnover’ conditions in which equal concentrations of protein and α-labeled ATP probe were incubated in the presence or absence of excess ssDNA. Probe binding was then measured by native polyacrylamide gel EMSA, while probe hydrolysis in identical samples was measured by TLC (Figure 5). All experiments were performed under salt conditions permissive for H352A catalytic activity. As shown in Figure 5A, the electrophoretic mobility of free α-labeled ATP (Figure 5A, lanes 1–2) is distinct from that of the protein-nucleotide complexes that form either in the absence (lanes 3–5) or presence (lanes 6–8) of ssDNA. The Rad51–ADP complex appears to dissociate from the ssDNA as suggested by the similar mobility shifts seen in the presence/absence of ssDNA (Figure 5A) and by previous studies (1,3,35). This allowed quantification of the nucleotide-binding data in Figure 5A by phosphorimaging, the results of which are shown in Figure 5B. In the absence of ssDNA, H352A has ∼3-fold weaker nucleotide-binding activity than wild-type, whereas H352Y binds nucleotide slightly better than wild-type (Figure 5B). ssDNA stimulates nucleotide binding >2.5-fold for wild-type and >5-fold for H352A, but has little effect on nucleotide binding by H352Y (Figure 5B). Even when stimulated by ssDNA, H352A-nucleotide binding remains weaker than wild-type (Figure 5B). Therefore H352A appears to bind nucleotide relatively weakly, but can still be stimulated by ssDNA, at least under low salt conditions. On the other hand nucleotide binding is relatively insensitive to ssDNA binding in H352Y.
Figure 5.
Binding and hydrolysis of stoichiometric ATP by Rad51 and mutants. ATP binding and hydrolysis was measured at a 1:1 concentration ratio of protein to ATP. Nucleotide binding was measured by polyacrylamide gel EMSA, and hydrolysis was measured by TLC, as described under ‘Materials and Methods’ section. (A) 10 µM wild-type or mutant Rad51, as indicated, was incubated with 10 µM α-[32P]-ATP (10 µCi/ml) for 2 h either in the absence (lanes 3–5) or presence (lanes 6–8) of 30 µM φX174 ssDNA. Control experiments lacking enzyme or lacking both enzyme and ssDNA are shown in lanes 1 and 2, respectively. Samples were electrophoresed on native 12% polyacrylamide gels to separate free from bound nucleotide, which were detected and quantified by phosphorimaging. Other buffer components and assay conditions are described under ‘Materials and Methods’ section. (B) The percentage of bound nucleotide in (A), was quantified for each form of Rad51 in the absence (gray bars) and presence (white bars) of ssDNA. (C) The percentage of ATP hydrolyzed in samples identical to those analyzed in (A), was measured for each form of Rad51 protein in the absence (gray bars) and presence (white bars) of ssDNA.
Figure 5C shows the percentage of ATP hydrolyzed during incubation of wild-type or mutant enzymes with stoichometric ATP. For wild-type Rad51, the percentage of ATP hydrolyzed (Figure 5C) closely parallels the percentage of nucleotide bound (Figure 5B) in both the absence and presence of ssDNA. This suggests that both free and ssDNA-bound Rad51 quantitatively hydrolyze bound ATP to ADP under these conditions, and subsequently remain in an ADP-bound state. Figure 5C also shows that hydrolysis of ATP by H352A is strongly dependent on ssDNA (13-fold stimulation) under single-turnover conditions; similar to what was observed in steady-state reactions under multiple-turnover (Vmax) conditions (Figure 4). Comparison of Figure 5B and C indicates that most of the ATP bound by H352A in the absence of ssDNA is not hydrolyzed, whereas most of the substrate is hydrolyzed in the presence of ssDNA. The single-turnover conditions may allow H352A to remain bound to ADP following catalysis (Figure 5B and C), although the mutant is clearly capable of multiple turnovers as demonstrated in Figure 4.
H352Y exhibits the highest level of ssDNA-independent ATP hydrolysis and is weakly stimulated by ssDNA (<1.5-fold) under single-turnover conditions (Figure 5C). Comparison of Figure 5B and C suggests that similar to wild-type Rad51, both free and ssDNA-bound H352Y quantitatively hydrolyze bound ATP to ADP and remain in the ADP-bound state thereafter. Taken together, the data in Figures 4 and 5 indicate that H352Y is proficient in catalyzing a single round of ATP hydrolysis, but deficient in performing multiple rounds of ATP hydrolysis.
Effects of His352 mutations on RPA displacement from ssDNA
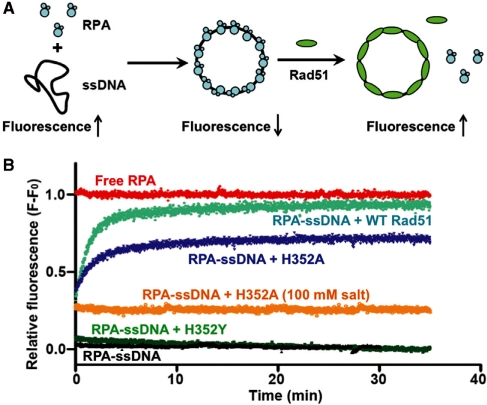
RPA has a high level of intrinsic tryptophan fluorescence that is quenched ∼40% upon binding to ssDNA, whereas Rad51 has no tryptophan residues and a low intrinsic fluorescence that is not quenched by ssDNA (21). These properties have been exploited to measure the displacement of RPA by Rad51 during presynaptic filament assembly, by following the increase in RPA tryptophan fluorescence upon addition of Rad51 to RPA–ssDNA (21). We used this technique to compare the abilities of wild-type and mutant Rad51 proteins to displace RPA protein from ssDNA (Figure 6). Control reactions demonstrate the maximum fluorescence signal for free RPA in red, and the minimum signal for intact RPA–ssDNA complex in black (Figure 6B). In the presence of ATP, the addition of wild-type Rad51 (Figure 6B, cyan) to pre-formed RPA–ssDNA causes a time-dependent increase in RPA fluorescence, consistent with published results (21,36). RPA displacement by wild-type Rad51 approaches 95% and is ∼90% complete within the first 5 min of the reaction (Figure 6B, cyan). The RPA fluorescence increase occurs with multi-phasic kinetics; i.e. there is a fast component of the reaction that is complete before signal acquisition occurs (dead time ≈5 s) that accounts for ∼25% of the total signal change. A similar fluorescence burst was observed in an earlier study (21). The H352A mutant (Figure 6B, blue) also displaces RPA from ssDNA at low salt and in the presence of ATP, i.e. conditions in which H352A is catalytically active. The reaction with H352A is slower than wild-type, and the total extent of RPA displacement only approaches 75% over 35 min. Like wild-type, the RPA fluorescence increase caused by H352A includes an unresolved fast component (Figure 6B, blue). Raising the salt concentration from 50 to 100 mM eliminates the slower component of RPA displacement by H352A so that only the unresolved component remains (Figure 6B, orange). Thus there is an immediate jump to ∼25% of the signal of free RPA upon addition of H352A, but the signal remains flat for 35 min thereafter. No fluorescence change occurs upon addition of Rad51 and ATP to RPA in the absence of ssDNA (data not shown). One possible interpretation of these results is that Rad51 rapidly forms an intermediate complex with RPA–ssDNA (partial recovery of fluorescence) before slowly displacing RPA from ssDNA (complete recovery of fluorescence). According to this scenario, with H352A the slow displacement phase is inhibited by salt while the formation of the intermediate complex is unaffected. An alternative interpretation is that there are different populations of RPA–ssDNA in the reactions, some of which are easily disrupted by Rad51 while others are more resistant. Further experimentation will be necessary to resolve between these and other models.
Figure 6.
RPA displacement assays. (A) The displacement of RPA protein from ssDNA by Rad51 wild-type and mutant proteins was monitored by the increase in RPA tryptophan fluorescence as described under ‘Materials and Methods’ section. (B) Continuous fluorescence assays for RPA displacement. All reactions contained 2.5 mM ATP, 0.25 µM RPA and 5 µM φX174 ssDNA, except for the ‘RPA alone’ control reaction (red) which lacked ssDNA. Reactions contained 2 µM of either wild-type (cyan), H352A (blue, orange), or H352Y (green) Rad51, except for the ‘RPA alone’ and ‘RPA–ssDNA’ (black) controls which lacked Rad51. All reactions contained 50 mM monovalent salt except for one reaction with H352A that contained 100 mM monovalent salt (orange). Other buffer components and reaction conditions are described under ‘Materials and Methods’ section.
The H352Y mutant (Figure 6B, green) does not remodel or displace RPA from ssDNA, since the fluorescence signal of RPA–ssDNA does not change upon addition of H352Y in the presence of ATP. Thus the intrinsically high ssDNA-binding affinity of H352Y is insufficient for RPA displacement from ssDNA.
Structure of the Rad51 H352Y mutant
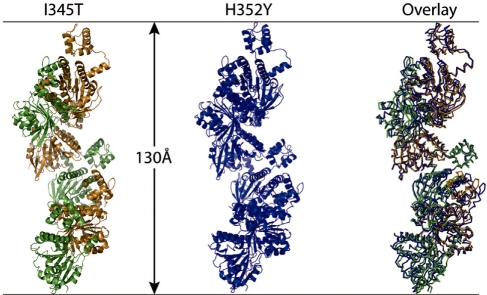
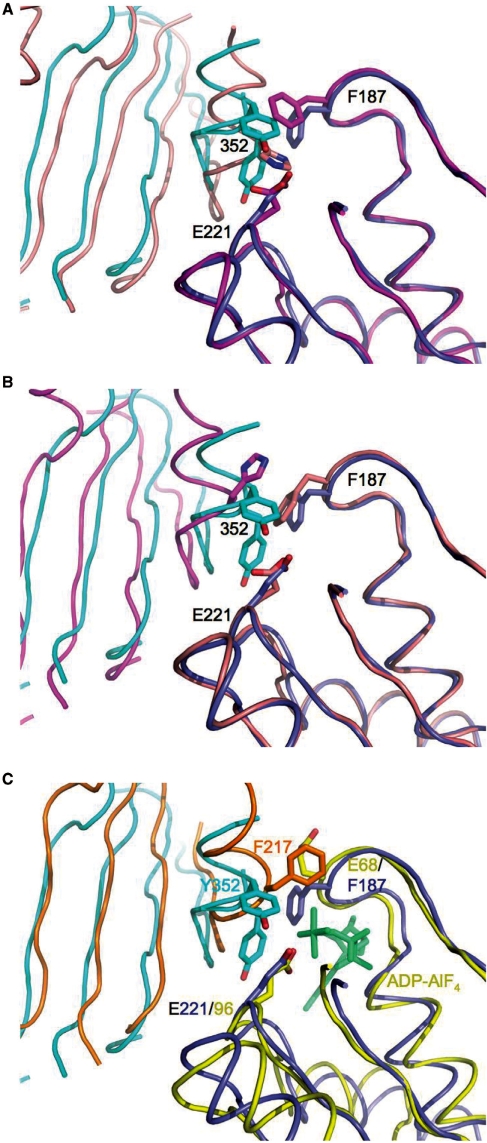
Like the majority of RecA-like proteins, Rad51 H352Y crystallizes as a filament with 61 helical symmetry. Figure 7 (center) shows one helical turn of the Rad51 H352Y filament containing six crystallographically identical subunits. The pitch of the filament, 130.3 Å, places it in the ‘high-pitch’ category for this family of proteins. For comparison, Figure 7 (left) shows the structure of the Rad51 I345T filament (11) (PDB code 1SZP), and the two filaments are superimposed on the right of Figure 7. The two filaments are remarkably similar in overall structure even though the I345T filament is composed of dimers with 31 helical symmetry. The protomer of Rad51 H352Y is also remarkably similar to the protomers of the I345T crystal structure, with RMS residuals after superimposing all C-α atoms of 1.10 and 1.21 Å (Figure 8A and B). The only significant variation in overall conformation between these protomers lies in the disposition of the 78–158 sub-domain, which rotates relative to the core of the protein. Omitting this sub-domain, which replaces the long N-terminal helix of the bacterial RecA proteins, the RMS residuals are cut nearly in half to 0.52 and 0.71 Å for the two protomers of the I345T dimer.
Figure 7.
The H352Y interface mutant of yeast Rad51 (center, blue) forms a 61 helical filament with a pitch of 130 Å. In comparison, the I345T mutant of this protein (left, green and gold, PDB code 1SZP) forms a filament of the same pitch but as a 31 helix of dimers. Despite the difference in helical symmetry, the filaments superimpose remarkably well (right.) Both proteins crystallized in the nucleotide-free state.
Figure 8.
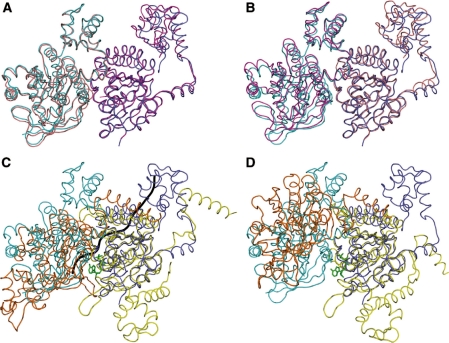
The protomers and the dimer interfaces of the yeast Rad51 H352Y, Rad51 I345T and E. coli RecA recombinases are remarkably similar. (A) The ATPase core of the ‘A’ protomer of Rad51 I345T (magenta, PDB code 1SZP) was superimposed onto a protomer of Rad51 H352Y (dark blue), carrying along the adjacent protomer ‘D’ (salmon for I345T, cyan for H352Y) but not including it in the superposition; (B) Likewise, superimposing only the ‘D’ protomer of I345T onto H352Y. (C) While the ATPase core of one protomer of the RecA–ssDNA-ADP.AlF4 complex (yellow, PDB code 3CMW) superimposes well onto that of Rad51-H352Y (dark blue), the adjacent protomer (not included in the superposition, orange for RecA, light blue for Rad51) overlays well only in the region of the dimer interface. The ADP-AlF4 (green) and ssDNA backbone trace (gray) of the RecA complex are shown for reference. (D) In contrast, the DNA-free form of E. coli RecA (PDB 3CMV) presents a substantially different dimer interface than the others.
The active site in Rad51/RecA proteins is formed by two adjacent protomers of the filament, one of which contains the nucleotide-binding pocket with Walker A (P-loop) and Walker B motifs, the other providing one wall of the cleft, with residue 352 located centrally on this latter surface. Since alternate protomers in the Rad51 I345T crystal structure are different, there are two different interfaces present in that filament. Both are very similar to the single protomer–protomer interface in H352Y: The ‘A–D’ dimer (named using the chain identifiers within the 1SZP PDB file, with the first chain being the one that presents its nucleotide-binding pocket at the dimer interface) of I345T superimposes with an RMS residual of 1.47 Å (Figure 8A), and the ‘D–A’ dimer superimposes with a residual of 1.05 Å (Figure 8B) (using all C-α atoms but excluding the 78–158 sub-domain in both cases). Interestingly, if the ‘A–D’ dimer of I345T is superimposed on the H352Y dimer using only the core (residues 159–396) of the ‘A’ protomer, the 78–158 sub-domain of the ‘D’ protomer (not used in the superposition) overlays remarkably well (0.72 Å RMS) on the corresponding sub-domain that bridges the dimer interface of H352Y (Figure 8A). When only the core of the ‘D’ protomer of the ‘D–A’ dimer is used, however, such a good overlap of the 78–158 sub-domains is not obtained (1.53 Å RMS) (Figure 8B).
The mutated residue, Tyr352, is seen to adopt two conformations in the H352Y crystal structure with approximately equal occupancy (Figure 9A and B). One of these is in nearly the same conformation as His352 of protomer D in the I345T structure; the second conformation of Tyr352 is substantially different from any of the others, involving changes to the backbone that allow the sidechain to lie almost parallel to the axis of the α-helix that precedes it. In the former conformation, Tyr352 forms a hydrogen bond to the carboxyl sidechain of Glu221 (2.6 Å) of the adjacent protomer, the putative nucleophilic activator for ATP hydrolysis. In the latter conformation, the tyrosine packs against Phe187 and Thr223 of the adjacent protomer. Other than a difference in rotamer for Phe187 and a concomitant shift in Arg188, sidechains in the active sites of the H352Y and I345T structures are essentially the same.
Figure 9.
Tyrosine 352 of the Rad51 H352Y mutant is present in two predominant alternate conformations. Detailed view of the dimer interfaces from the perspective of the ssDNA, using the superpositions as described in Figure 8, with the protomers on the right half of each figure used in the superposition. (A) One of the alternate conformations of Tyr352 of H352Y (cyan) is similar to His352 of the ‘D’ protomer (salmon) in the ‘AD’ dimer of I345T. (B) Neither conformation of Tyr352 (cyan) is similar to His352 of the ‘A’ protomer (magenta) of the ‘DA’ dimer of I345T. (C) While there is a good match between the dimer interfaces of Rad51 H352Y (dark blue and cyan) and RecA–ssDNA–ADP–AlF4 (yellow and orange), Phe217, equivalent to residue 352 of yeast, is disposed differently than either alternate conformation of Tyr352.
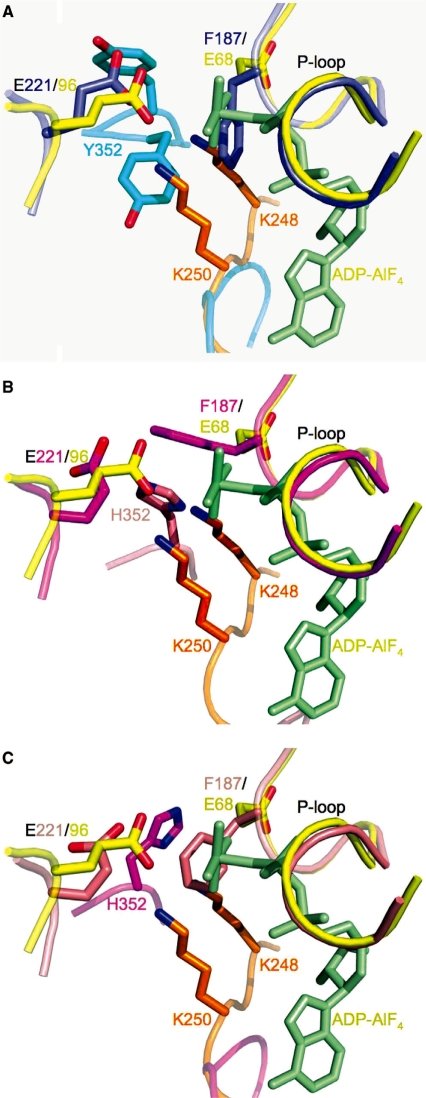
Surprisingly, the dimers comprising the active site of Rad51 superimpose remarkably well on dimers of the E. coli RecA–DNA crystal structures (4) (Figures 8C and 9C), with an RMS residual of 2.13 Å for 317 C-α atoms of the RecA–ssDNA–ADP–AlF4 complex (PDB code 3CMW.) In contrast, the Rad51 dimers do not superimpose well on dimers of the DNA-free form of E. coli RecA (4) (Figure 8D). The Walker A/B motifs are in nearly an identical overall conformation, with an RMS residual of 0.58 Å for the 13 C-α atoms, despite the presence of a bound ADP-AlF4 in the RecA–ssDNA structure and only a chloride ion in the P-loop of the Rad51 H352Y structure (Figure 10A). The sidechain of the putative catalytic glutamate (Glu221 in Rad51, Glu96 in RecA) is located similarly in these structures. Sidechains of some key residues in the active site region are significantly different, however, with potential mechanistic implications. The conformation of Phe187 in Rad51 H352Y, which is in the middle of the Walker A (P-loop) motif, sterically overlaps with the AlF4 moiety that mimics the γ-phosphate of an ATP in the RecA–ssDNA–ADP–AlF4 complex (Figure 10A and Supplementary Figure 2). A similar conformation of Phe187 and subsequent steric overlap also occurs in protomer D of Rad51 I345T (Figure 10C); in protomer A, Phe187 is swung out but still sterically excludes the γ-phosphate (Figure 10B). The equivalent residue in RecA, Glu68, points away from the P-loop pocket.
Figure 10.
Superposition of the Walker A and B motifs of E. coli RecA–ssDNA–ADP–AlF4 onto the Rad51–H352Y and -I345T mutants suggests a key role for P-loop residue Phe187 (Glu68 in E. coli) in sensing the gamma-phosphate or preventing the binding of ATP, and for residue 352 in assisting in catalysis. RecA is shown in yellow with its adjacent protomer in orange and its bound ADP-AlF4 in light green in these figures. (A) Superimposed Rad51-H352Y (purple) and its adacent protomer (cyan); (B) Superimposed ‘A’ protomer of Rad51–I345T (magenta) with its adjacent ‘D’ protomer (salmon); (C) Superimposed ‘D’ protomer of Rad51–I345T (salmon) with its adjacent protomer (magenta.) In (A) and (C), Phe187 of the P-loop is only 1.65 Å and 1.70 Å, respectively, from the AlF4 mimic of the γ-phosphate of the ATP. In (B), the δ-nitrogen of His352 is seen to be located almost exactly in between the amine groups of lysines 248 and 250 of E. coli, residues which have been shown to be essential for nucleotide hydrolysis.
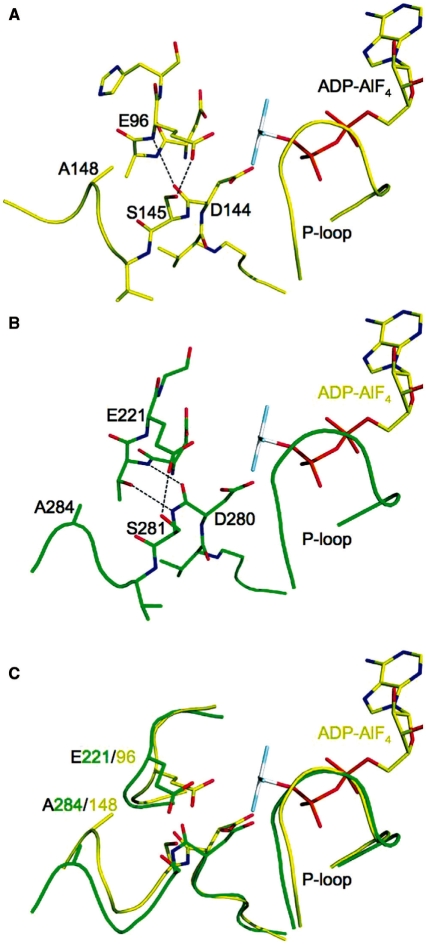
At the end of the Walker B motif, the peptide linkage between the highly conserved Asp280 and Ser281 residues is in the relatively rare cis conformation (i.e. with both the backbone carbonyl oxygen and amide hydrogen on the same side of the amide linkage) in both the Rad51 H352Y (Figure 11B) and I345T structures, as it is in nearly all crystal structures of RecA family proteins except those which have DNA bound. In the cis conformation (as represented by the Rad51 H352Y structure in Figure 11B), the backbone amide NH of the cis peptide linkage hydrogen bonds to the hydroxyl sidechain of Thr220, the residue preceding the putative catalytic Glu221. No such bond is formed involving the trans backbone amide NH in the RecA–DNA complexes (represented by 3CMW, with similar interactions in the other RecA–DNA complexes) (Figure 11A, and shown in superposition with Rad51 H352Y in Figure 11C). Interactions of the backbone carbonyl of this linkage differ markedly between the cis and trans states: In Rad51, the cis carbonyl hydrogen bonds to the backbone amide of residue 220, whereas in the RecA–DNA complex it forms a hydrogen bond to the backbone amide of Glu96 (the putative catalytic residue in RecA.) Ser281/145 undergoes a major conformational rearrangement in going from the cis to trans state: In both states it bonds to the carboxyl of the catalytic glutamate, but in the cis state the serine is shifted away from the P-loop, thus pulling the carboxyl group away from the active site (Figure 11A and C); whereas in the trans state in the RecA–DNA complex, the equivalent Ser145 is pushed towards the P-loop and makes very close contact (2.42 Å) with the carboxyl group of Glu96, resulting in a shift of this putative nucleophilic activator toward the γ-phosphate of the ATP (Figure 11B and C). Furthermore, as a consequence of the backbone geometry imposed by the cis/trans linkage, residues 146–148 in the trans state in RecA–DNA are shifted toward the P-loop, placing Ala148 in close VDW contact with the Cγ (4.19 Å) and α-carbon (3.84 Å) of Glu96 (Figure 11B and C); whereas in the cis state represented by Rad51, the corresponding residues 282–284 are again shifted away from the P-loop, and Ala284 is seen to be no closer than 5.7 Å to Glu221 (Figure 11A and C). The net difference in position of the putative catalytic glutamate sidechain is a shift of 1.25 Å towards the γ-phosphate of the ATP in the trans peptide state represented by the DNA-bound RecA complex relative to the cis peptide state as seen in the DNA-free Rad51 states (of either the H352Y or I345T structures.).
Figure 11.
A role for cis to trans isomerization of the peptide linkage at the end of the conserved Walker B motif in configuring the active site for nucleotide hydrolysis. (A) The only RecA-family structures to clearly possess a trans peptide linkage between residues 144–145 at the end of Walker B contain DNA, as represented here by the active site of E. coli RecA–ssDNA–ADP–AlF4 complex (PDB code 3CMW). (B) As in nearly all other RecA-family structures, the active site of Rad51–H352Y shows a cis configuration at this peptide (between residues 280–281). (C) Compared with the cis configuration, in the trans configuration Ser281 (145 in E. coli) and Ala284 (148 in E. coli) move toward the active site, and push the putative nucleophilic activator Glu221 (96 in E. coli) 1.25 Å closer to the γ-phosphate (mimicked by the AlF4 moiety) of the ATP.
DISCUSSION
Role of His352 in Rad51 ssDNA-binding and catalytic activities
The biochemical data demonstrate that residue His352 at the filament interface plays a critical role in modulating the affinity and/or cooperativity of Rad51–ssDNA interactions, and in the coupling of ssDNA binding to catalysis. Mutation of His352 is detrimental to strand exchange activity under the relatively stringent conditions imposed by long DNA substrates and by the presence of RPA. In addition, the opposite effects of the alanine and tyrosine substitutions on ssDNA-binding activity indicate different mechanisms for the strand exchange defects of Rad51 H352A and H352Y.
The H352A mutant appears to be a classic weak ssDNA binder, based on the increased salt-sensitivity of H352A–ssDNA interactions compared with wild-type Rad51. The instability of H352A–ssDNA interactions could be a result of lower intrinsic affinity for ssDNA, lower cooperativity (e.g. weak protomer–protomer interactions), or both. Fundamentally, H352A is catalytically competent. At low salt concentrations H352A catalyzes a robust ssDNA-dependent ATPase reaction (Figure 4), consistent with results of a previous study (13), and it displaces RPA from ssDNA, albeit at a slower rate and efficiency than wild-type (Figure 6). Both ATPase and RPA displacement activities are suppressed by small increases in salt concentration that weaken H352A–ssDNA interactions (Figures 3, 4 and 6). H352A exhibits reduced DNA strand exchange activity compared with wild-type in reactions using either long DNA substrates in the presence of RPA (Figure 1) or short oligonucleotide substrates in the absence of RPA (13). These findings suggest that the DNA strand exchange defect of H352A is due to the formation of relatively unstable filaments on ssDNA.
In contrast, the H352Y mutant is a constitutively strong ssDNA binder (Figure 3), but it is incapable of DNA strand exchange using either long DNA substrates in the presence of RPA (Figure 1) or short oligonucleotide substrates in the absence of RPA (13). H352Y is capable of ATP hydrolysis, but this activity is effectively limited to a single turnover (Figures 4 and 5), indicating a defect in the exchange of ADP product for a new ATP substrate molecule. This nucleotide exchange defect of H352Y is observed both in the absence and in the presence of ssDNA. The data indicate that nucleotide exchange is important for the DNA strand exchange activity of Rad51.
Despite its constitutively strong ssDNA-binding activity (Figure 3), H352Y fails to displace RPA from ssDNA in the presence of ATP (Figure 6). This finding demonstrates that tight ssDNA binding alone is insufficient for RPA displacement. Rather, the data indicate that ssDNA binding by Rad51 must be coupled to ATP hydrolysis and nucleotide exchange in order for RPA displacement to occur. This is consistent with the observation that the alanine mutant does promote RPA displacement under salt conditions that are permissive for multiple turnovers of its ssDNA-dependent ATPase activity. Although our RPA displacement and DNA strand exchange assays used opposite orders of protein addition to ssDNA, it is possible that the inability of H352Y to displace RPA from ssDNA may be a contributing factor to its inability to promote DNA strand exchange reactions on long DNA substrates. The data suggest that the coupling of ssDNA binding to ATP hydrolysis (or to a conformational change to a hydrolysis-competent form) is essential for Rad51 to displace RPA from ssDNA. This coupling is achieved by the H352A mutant under permissive salt conditions, despite its lower ssDNA-binding affinity, but not by H352Y.
Structure of the Rad51-H352Y mutant
The structure of the Rad51 H352Y mutant is remarkably similar overall and in detail to the I345T mutant (11), suggesting that the I345T and H352Y mutations within the two crystallized proteins do not affect the overall form or pitch of the DNA-free Rad51 filament (or coincidentally do so to the same extent.) The yeast-specific N-terminal extension (residues 1–79), which is present in our protein but disordered within the crystal, and which was deleted in order to obtain crystals of I345T, likewise does not affect the structure of the DNA-free, nucleotide-free Rad51 filament. Both filaments are in the ‘high-pitch’ form, generally regarded as the active form (5). The main difference between the two structures is that the H352Y filament exhibits 61 symmetry with the protomer as the repeating unit, while the I345T filament exhibits 31 symmetry with the dimer as the repeating unit (11). As a result, I345T filaments contain two different types of protomer–protomer interfaces and two different types of active sites, involving different interactions of His352 with the active site and with Phe187 from the adjacent subunit. In contrast the H352Y filament contains one type of interface, one type of active site, and a single geometry for Phe187.
It is interesting to note that the crystal structure of the I345T mutant actually contains six independent protomers arranged as three distinct filaments (11). While the A–D and F–E filaments are essentially identical, the third filament, B–C, is nearly a 61 helix like that in the H352Y filament, and also shows conformational disorder in the helix that contains His352 and in the Phe187 sidechain. The alternating dimers in the A–D and F–E filaments, with Phe187 alternately projecting into and away from the γ-phosphate-binding region of the Walker A motif, along with the multiple conformations seen for Tyr352 (which interacts with Phe187) in our structure, suggests that the active site configurations represented by protomers A, D, E and F in the I345T structure may represent stable intermediates along the recombination pathway, and that our structure and the B and C protomers of the I345T structure are transient intermediates between those two structures.
Role of Phe187 in Rad51 mechanism
Our structure in comparison with other RecA family proteins supports a key role for Phe187 in the recombinase pathway. Crystallized Rad51 filaments (H352Y and I345T) are captured in a conformation that does not allow binding of the triphospho nucleotide: This is supported by the fact that no nucleotide was seen in the I345T crystal structure, in spite of the presence of 10 mM ATPγS in the crystallization mixture (11). Superimposing the RecA–DNA–ADP–AlF4 structure on the Rad51 structures shows that Phe187 sterically excludes the γ-phosphate. The I354T filament structure must represent a transient intermediate, since the mutant is strand exchange-competent. This suggests that in the Rad51 mechanism, Phe187 could either serve as a sensor of the nucleotide state, forced out of the nucleotide-binding pocket by the presence of ATP, or as a regulator of ATP binding, preventing binding of ATP by steric exclusion of the γ-phosphate. In the case of H352Y, however, the filament becomes trapped in this conformation after hydrolyzing 1 ATP, resulting in an inability to exchange the nucleotide. This is a potential structural explanation for the greatly reduced hydrolytic activity of the H352Y mutant. By virtue of its interactions with residue 352 of the adjacent protomer comprising the active site, the conformation of Phe187 appears to be dependent on the configuration of the protomer–protomer interface, or conversely, that the configuration of the interface depends on the conformation of Phe187. In either case, this provides a structural mechanism for the roles that Phe187 (Glu68 in E. coli RecA) and His352 (Phe217 in RecA) have been observed to play in communication between the active site and the state of the filament interface. Given the similarity of the dimer interface observed in the E. coli RecA–ssDNA–ADP–AlF4 crystal structure with that seen in both S. cerevisiae Rad51 crystal structures, it is tempting to speculate that after hydrolysis of the ATP, Glu68 of the Walker A motif, equivalent to Phe187 in Rad51, forms a salt bridge to either or both of Lys248 and Lys250 that sterically blocks nucleotide exchange and holds the filament in this conformation until an appropriate ‘signal’ is obtained.
Role of cis/trans peptide linkage in signaling the DNA-bound state
In comparison with the RecA–DNA structures (4), our structure provides a possible role for the cis/trans peptide linkage at the end of the Walker B motif (Figure 11). In virtually all RecA family structures except those that contain DNA, this peptide linkage (e.g. between residues 280 and 281 in Rad51, 144 and 145 in E. coli RecA) is in the relatively rare cis conformation (4,11,37). In this conformation, the putative catalytic glutamate (Glu221 in Rad51, 96 in E. coli RecA) is pulled away from the γ-phosphate (as modeled by the ADP–AlF4 analog in the RecA–DNA structures) by Ser281/145 and backbone interactions between the cis peptide and the backbone and sidechain of Thr220. In contrast, the trans linkage enforces three structural changes in its vicinity that result in a shift of Glu221/96 towards the γ-phosphate and that greatly reduce that sidechain’s degrees of freedom. The trans linkage induces a substantial rearrangement of Ser281/145, placing it in close steric contact with the carboxylate of Glu221/96 while maintaining reasonable H-bonding geometry. The carbonyl of the trans peptide bonds with the backbone of Glu221/96, further locking that residue in place. And thirdly, as a consequence of the trans peptide, Ala284/148 pushes the sidechain of Glu221/96 closer to the γ phosphate. The trans linkage thus appears to orient and lock the glutamate residue in its catalytically competent conformation for ATP hydrolysis.
It does not appear that the cis/trans peptide linkage is dictated by the nucleotide state, since the cis state is observed in virtually all structures of RecA family proteins, whether in the apo, ADP-bound, or ATP-analog bound forms (4,11,37). Occurrence of the trans peptide linkage correlates only with the presence of the ssDNA or dsDNA. The propagation pathway is not clear from the structures, but it is noteworthy that DNA-binding loop L1 (289–296/157–164) follows the Walker B motif, and that DNA-binding loop L2 (327–344/195–209) follows the ‘sensor residue’ Gln326/194 and extends from the beta-strand adjacent to the Walker A motif. Indeed, across the RecA family, there is significant variation in the sequence and structure of the protein from residue 283/147 through the disordered loop. Thus, while the mechanism for conveying the signal remains to be determined, it appears that the presence of DNA is responsible for the cis to trans state change that ultimately results in pushing the putative catalytic glutamate closer to the γ phosphate and holding it in place to activate a water molecule for nucleophilic attack.
ACCESSION NUMBER
PDB ID 3LDA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Cancer Institute, NIH, HHS, USA (Program Project Grant no. P01 CA098993); NCI (Y1-CO-1020 to GM/CA-CAT); NIGMS, NIH, HHS, USA (Y1-GM-1104); US Department of Energy, Basic Energy Sciences, Office of Science (under contract No. DE-AC02-06CH11357 for the use of Advanced Photon Source). Funding for open access charge: National Institutes of Health (grant no. P01 CA098993).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs Tom Ellenberger and Marc Wold, respectively, for providing us with expression plasmids for the yeast Rad51 and RPA proteins. We thank Pierre Aller and Karl Zahn for data collection at Advanced Photon Source synchrotron.
REFERENCES
- 1.Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci. 1998;3:D570–D603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 2.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 3.Chi P, Van Komen S, Sehorn MG, Sigurdsson S, Sung P. Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair. 2006;5:381–391. doi: 10.1016/j.dnarep.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–484. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 5.Sheridan SD, Yu X, Roth R, Heuser JE, Sehorn MG, Sung P, Egelman EH, Bishop DK. A comparative analysis of Dmc1 and Rad51 nucleoprotein filaments. Nucleic Acids Res. 2008;36:4057–4066. doi: 10.1093/nar/gkn352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 7.Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- 8.Beernink HT, Morrical SW. The uvsY recombination protein of bacteriophage T4 forms hexamers in the presence and absence of single-stranded DNA. Biochemistry. 1998;37:5673–5681. doi: 10.1021/bi9800956. [DOI] [PubMed] [Google Scholar]
- 9.Fortin GS, Symington LS. Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing the stability of Rad51-DNA complexes. EMBO J. 2002;21:3160–3170. doi: 10.1093/emboj/cdf293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelley JA, Knight KL. Allosteric regulation of RecA protein function is mediated by Gln194. J. Biol. Chem. 1997;272:25778–25782. doi: 10.1074/jbc.272.41.25778. [DOI] [PubMed] [Google Scholar]
- 11.Conway AB, Lynch TW, Zhang Y, Fortin GS, Fung CW, Symington LS, Rice PA. Crystal structure of a Rad51 filament. Nat. Struct. Mol. Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 12.De Zutter JK, Knight KL. The hRad51 and RecA proteins show significant differences in cooperative binding to single-stranded DNA. J. Mol. Biol. 1999;293:769–780. doi: 10.1006/jmbi.1999.3200. [DOI] [PubMed] [Google Scholar]
- 13.Grigorescu AA, Vissers JH, Ristic D, Pigli YZ, Lynch TW, Wyman C, Rice PA. Inter-subunit interactions that coordinate Rad51′s activities. Nucleic Acids Res. 2009;37:557–567. doi: 10.1093/nar/gkn973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lefebvre SD, Morrical SW. Interactions of the bacteriophage T4 gene 59 protein with single-stranded polynucleotides: binding parameters and ion effects. J. Mol. Biol. 1997;272:312–326. doi: 10.1006/jmbi.1997.1264. [DOI] [PubMed] [Google Scholar]
- 15.Menetski JP, Kowalczykowski SC. Interaction of recA protein with single-stranded DNA. Quantitative aspects of binding affinity modulation by nucleotide cofactors. J. Mol. Biol. 1985;181:281–295. doi: 10.1016/0022-2836(85)90092-0. [DOI] [PubMed] [Google Scholar]
- 16.Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 17.Zaitseva EM, Zaitsev EN, Kowalczykowski SC. The DNA binding properties of Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 1999;274:2907–2915. doi: 10.1074/jbc.274.5.2907. [DOI] [PubMed] [Google Scholar]
- 18.Sweezy MA, Morrical SW. Single-stranded DNA binding properties of the uvsY recombination protein of bacteriophage T4. J. Mol. Biol. 1997;266:927–938. doi: 10.1006/jmbi.1996.0829. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, Qian N, Morrical SW. Dynamics of bacteriophage T4 presynaptic filament assembly from extrinsic fluorescence measurements of Gp32-single-stranded DNA interactions. J. Biol. Chem. 2006;281:26308–26319. doi: 10.1074/jbc.M604349200. [DOI] [PubMed] [Google Scholar]
- 20.Rice KP, Eggler AL, Sung P, Cox MM. DNA pairing and strand exchange by the Escherichia coli RecA and yeast Rad51 proteins without ATP hydrolysis: on the importance of not getting stuck. J. Biol. Chem. 2001;276:38570–38581. doi: 10.1074/jbc.M105678200. [DOI] [PubMed] [Google Scholar]
- 21.Sugiyama T, Kowalczykowski SC. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 2002;277:31663–31672. doi: 10.1074/jbc.M203494200. [DOI] [PubMed] [Google Scholar]
- 22.Ando RA, Morrical SW. Single-stranded DNA binding properties of the UvsX recombinase of bacteriophage T4: binding parameters and effects of nucleotides. J. Mol. Biol. 1998;283:785–796. doi: 10.1006/jmbi.1998.2124. [DOI] [PubMed] [Google Scholar]
- 23.Birdsall B, King RW, Wheeler MR, Lewis C.A., Jr, Goode SR, Dunlap RB, Roberts GC. Correction for light absorption in fluorescence studies of protein-ligand interactions. Anal. Biochem. 1983;132:353–361. doi: 10.1016/0003-2697(83)90020-9. [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.Kissinger CR, Gehlhaar DK, Fogel DB. Rapid automated molecular replacement by evolutionary search. Acta Crystallogr. D Biol. Crystallogr. 1999;55:484–491. doi: 10.1107/s0907444998012517. [DOI] [PubMed] [Google Scholar]
- 26.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 28.Leonard NJ. Etheno-substituted nucleotides and coenzymes: fluorescence and biological activity. CRC Crit. Rev. Biochem. 1984;15:125–199. doi: 10.3109/10409238409102299. [DOI] [PubMed] [Google Scholar]
- 29.Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 30.Hegner M, Smith SB, Bustamante C. Polymerization and mechanical properties of single RecA-DNA filaments. Proc. Natl Acad. Sci. USA. 1999;96:10109–10114. doi: 10.1073/pnas.96.18.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Noort J, Verbrugge S, Goosen N, Dekker C, Dame RT. Dual architectural roles of HU: formation of flexible hinges and rigid filaments. Proc. Natl Acad. Sci. USA. 2004;101:6969–6974. doi: 10.1073/pnas.0308230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim JM, Maraboeuf F, Kim SK, Shinohara A, Takahashi M. Effect of ions and nucleotides on the interactions of yeast Rad51 protein with single-stranded oligonucleotides. J. Biochem. 2001;129:469–475. doi: 10.1093/oxfordjournals.jbchem.a002879. [DOI] [PubMed] [Google Scholar]
- 33.Kiianitsa K, Solinger JA, Heyer WD. NADH-coupled microplate photometric assay for kinetic studies of ATP-hydrolyzing enzymes with low and high specific activities. Anal. Biochem. 2003;321:266–271. doi: 10.1016/s0003-2697(03)00461-5. [DOI] [PubMed] [Google Scholar]
- 34.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 35.van Mameren J, Modesti M, Kanaar R, Wyman C, Peterman EJ, Wuite GJ. Counting RAD51 proteins disassembling from nucleoprotein filaments under tension. Nature. 2009;457:745–748. doi: 10.1038/nature07581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kantake N, Sugiyama T, Kolodner RD, Kowalczykowski SC. The recombination-deficient mutant RPA (rfa1-t11) is displaced slowly from single-stranded DNA by Rad51 protein. J. Biol. Chem. 2003;278:23410–23417. doi: 10.1074/jbc.M302995200. [DOI] [PubMed] [Google Scholar]
- 37.McRobbie AM, Carter LG, Kerou M, Liu H, McMahon SA, Johnson KA, Oke M, Naismith JH, White MF. Structural and functional characterisation of a conserved archaeal RadA paralog with antirecombinase activity. J. Mol. Biol. 2009;389:661–673. doi: 10.1016/j.jmb.2009.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brunger AT. Free R value: cross-validation in crystallography. Methods Enzymol. 1997;277:366. doi: 10.1016/s0076-6879(97)77021-6. [DOI] [PubMed] [Google Scholar]
- 39.Luzzati V. Traitement statistique des erreurs dans la determination des structures cristallines. Acta Crystallogr. 1952;5:802–810. [Google Scholar]
- 40.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 41.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 42.Sugiyama T, Zaitseva EM, Kowaczykowski SC. A single-stranded DNA binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J. Biol. Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.