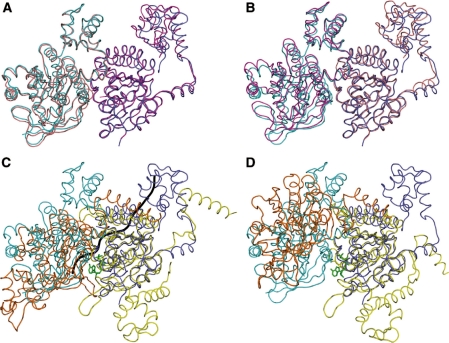
Figure 8.
The protomers and the dimer interfaces of the yeast Rad51 H352Y, Rad51 I345T and E. coli RecA recombinases are remarkably similar. (A) The ATPase core of the ‘A’ protomer of Rad51 I345T (magenta, PDB code 1SZP) was superimposed onto a protomer of Rad51 H352Y (dark blue), carrying along the adjacent protomer ‘D’ (salmon for I345T, cyan for H352Y) but not including it in the superposition; (B) Likewise, superimposing only the ‘D’ protomer of I345T onto H352Y. (C) While the ATPase core of one protomer of the RecA–ssDNA-ADP.AlF4 complex (yellow, PDB code 3CMW) superimposes well onto that of Rad51-H352Y (dark blue), the adjacent protomer (not included in the superposition, orange for RecA, light blue for Rad51) overlays well only in the region of the dimer interface. The ADP-AlF4 (green) and ssDNA backbone trace (gray) of the RecA complex are shown for reference. (D) In contrast, the DNA-free form of E. coli RecA (PDB 3CMV) presents a substantially different dimer interface than the others.