Figure 2.
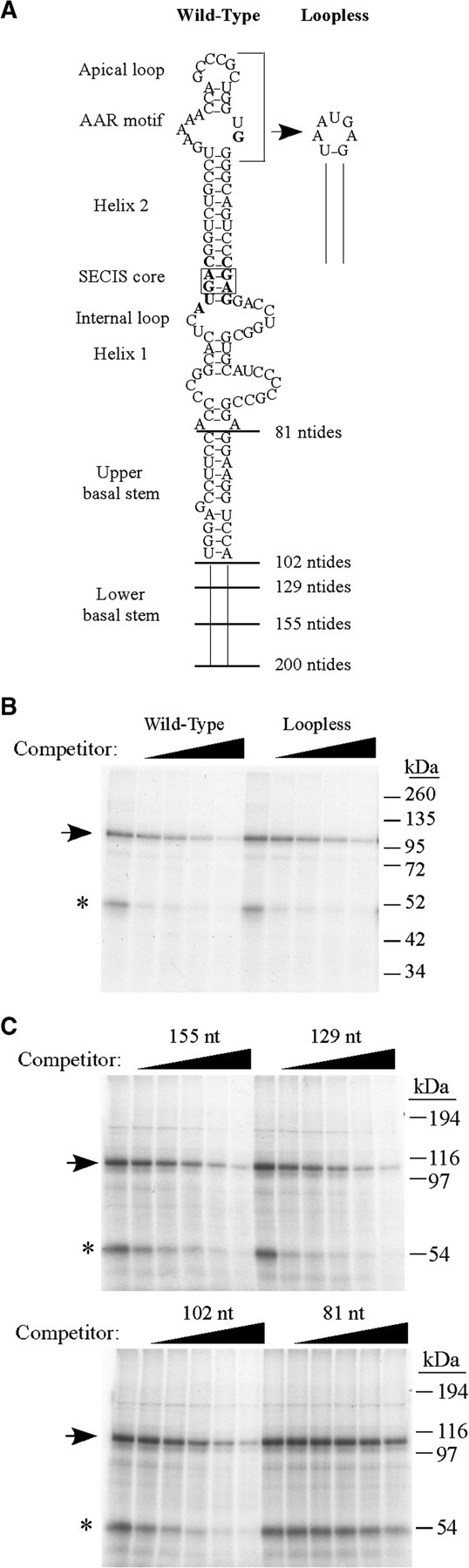
Analysis of mutant PHGPx SECIS RNAs to define the binding site requirements for the 110 kDa protein. (A) Schematic of the PHGPx wild-type SECIS (200 nt), the loopless mutant RNA, and the basal stem deletion mutants of 155, 129, 102 or 81 nt. (B) Nuclear extract from McArdle 7777 cells (2 µg) was preincubated with buffer or increasing amounts (0–200-fold molar excess over the probe) of unlabeled wild-type or loopless PHGPx SECIS RNAs for 10 min prior to the addition of the 32P-labeled PHGPx SECIS probe. The 110 kDa crosslinked product is indicated by the arrow and the asterisk indicates the 50 kDa protein of unknown identity. (C) Competition experiments were performed as in (B) using 5.5 µg nuclear extract and increasing amounts (0–500-fold molar excess) of cold basal stem deletion mutant RNAs of 155, 129, 102 and 81 nt, as indicated.
