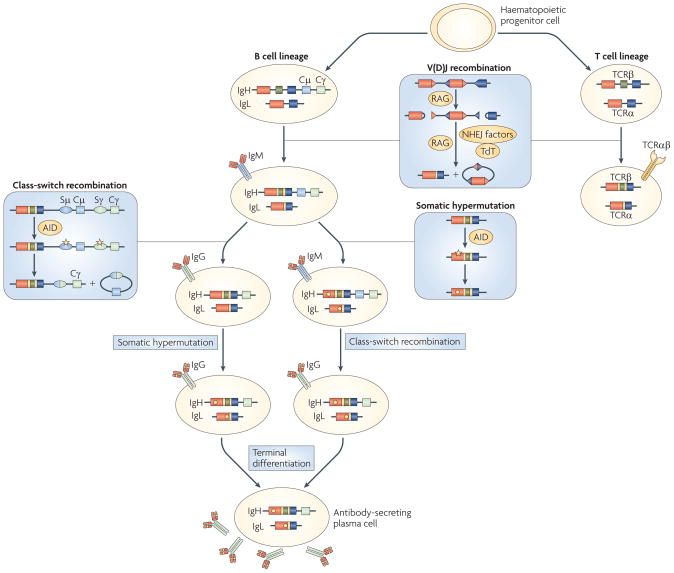
Figure 1. Lymphocyte development and antigen receptor diversification in jawed vertebrates.
A haematopoietic progenitor cell gives rise to distinct B and T cell lineages. Transcriptional networks (not depicted) are crucial for the differentiation and maintenance of cellular identity. Three unique processes — variable, diversity and joining region (V(D)J) recombination, somatic hypermutation and class-switch recombination — diversify antigen receptor genes. For clarity, some details are simplified or omitted. V (red boxes), D (green boxes) and J (dark blue boxes) segments for representative T cells (T cell receptor α-chain (TCRα) and TCRβ)) and B cells (immunoglobulin heavy chain (IgH) and immunoglobulin light chain (IgL)) are shown. The constant region for the Igμ isotype (Cμ) and a single representative downstream Cγ exon within the IgH locus are depicted. Key factors that facilitate each diversification step are shown in yellow ovals. During V(D)J recombination, recombination signal sequences (RSSs; blue and red triangles) direct the recombination-activating gene 1 (RAG1)–RAG2 recombinase complex to individual gene segments (red and blue boxes). The recombinase introduces two double-strand DNA breaks with blunt signal ends and hairpin-sealed coding ends. In the subsequent joining phase, terminal deoxynucleotidyltransferase (TdT), a template-independent DNA polymerase, adds random nucleotides to the junction of the gene elements, thereby increasing repertoire diversity dramatically; the RSSs are joined without further end processing and form excision circles. Once functional DNA rearrangements occur, TCR sequences are unaltered. After encounter with antigen, B cells further recombine the receptor by somatic hypermutation and class-switch recombination. Somatic hypermutation is initiated by activation-induced cytidine deaminase (AID), which deaminates individual cytidines within the V(D)J exon of the immunoglobulin gene, leading to U:G mismatches (yellow star). Subsequent error-prone repair results in individual point mutations (yellow dot in the gene and yellow bar in the immunoglobulin molecules), and B cells with higher affinity for the original antigen are selected. During class-switch recombination, AID creates U:G mismatches in the highly repetitive switch (S) regions (blue and green ovals) that are upstream of the exons encoding the constant regions of different isotypes. Error-prone repair leads to the generation of double-strand DNA breaks, excision of the intervening DNA (containing the Cμ exons) and joining of the remains of the switch regions. The recombined, somatically mutated V(D)J region is then associated with Cγ (green box), instead of Cμ. The precise order in which somatic hypermutation and class-switch recombination occur is unclear. Class-switch recombination has occurred in almost all high-affinity immunoglobulin-expressing B cells. In the course of a humoral immune response, B cells undergo terminal differentiation into plasma cells, which secrete large amounts of soluble immunoglobulins. γδ T cells — in which γδ TCR genes, instead of their αβ TCR genes, have been rearranged — and immunoglobulin gene conversion, which has been demonstrated in differentiating B cells of birds and rabbits, are not shown.