Fig. 1. Classification of viruses present in VLP preparations generated from fecal samples collected from four families of MZ twins and their mothers.
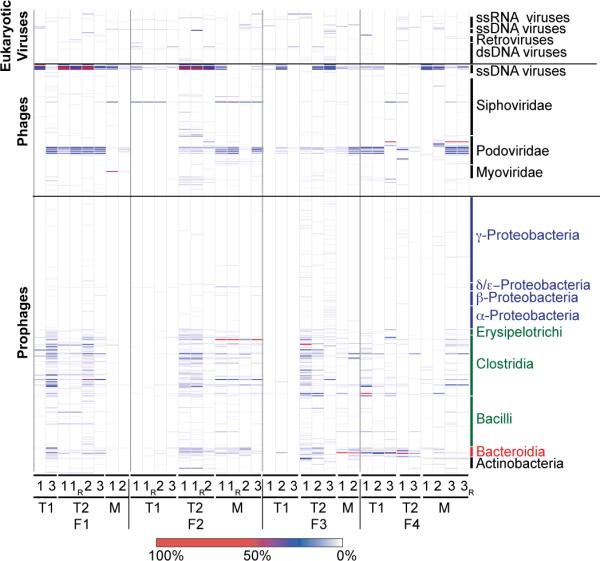
Prophages are classified based on their bacterial host taxonomy. Prominent bacterial phyla are represented by different colors (Proteobacteria, blue; Firmicutes green; Bacteroidetes, red; Actinobacteria, black). Class-level taxa within these phyla are indicated. Phage and eukaryotic viruses are sorted according to taxonomy. Nomenclature used for VLP preparations from fecal biospecimens: F, family; T1, co-twin 1; T2, co-twin 2; M, mother of co-twins. Time points (1–3), and technical replicates (R) produced from a given sample are noted. The color bar at the bottom of the figure provides a reference key for the percent coverage of a viral genome in the NR_Viral_DB by reads from given VLP virome dataset (data normalized using 14,000 randomly selected reads/dataset).
