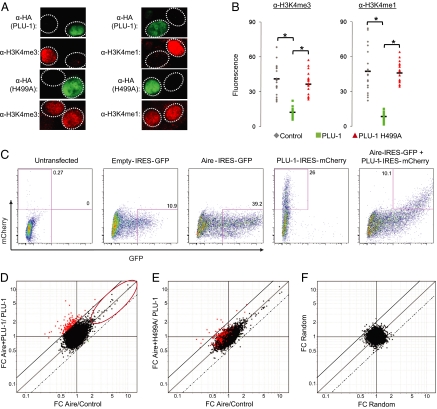
Fig. 4.
Broad H3K4 hypomethylation does not have a major effect on the range of Aire-targeted genes (A) Global H3K4 hypomethylation: Expression plasmids encoding HA-tagged PLU-1 or PLU-1 H499A were cotransfected with Aire on U2OS cells, and H3K4me1, H3K4m3 (both red), and HA (green) levels were analyzed via immunofluorescence. (B) Quantification for n = 20 cells by calculating mean fluorescence intensity of an invariant area. Filled bars indicate average mean intensity. *χ2 P value <10−4. (C) Bicistronic vectors were constructed with PLU-1, PLU-1H499A and/or Aire amino-terminal to an internal ribosome entry site driving mCherry or GFP. U2OS cells were transfected with an empty IRES-GFP vector, PLU-1-IRES-mCherry, Aire-IRES-GFP, or were cotransfected with Aire-IRES-GFP and PLU-1- or PLU-1H499A-IRES mCherry and sorted via a FACSAria. (D) FC scatterplots comparing effect of Aire in presence (y axis) and absence (x axis) of PLU-1. Red dots indicate genes differentially induced by Aire in the presence of PLU-1 vs. Aire alone. Red ellipse indicates genes induced by Aire alone (FC > 2). Solid diagonal above x = y indicates FC ratio of 2. Dotted diagonal below x = y indicates FC ratio of 0.5. (E) Same as D, but with PLU-1-H499A. (F) Same as D, but drawn from a random dataset.