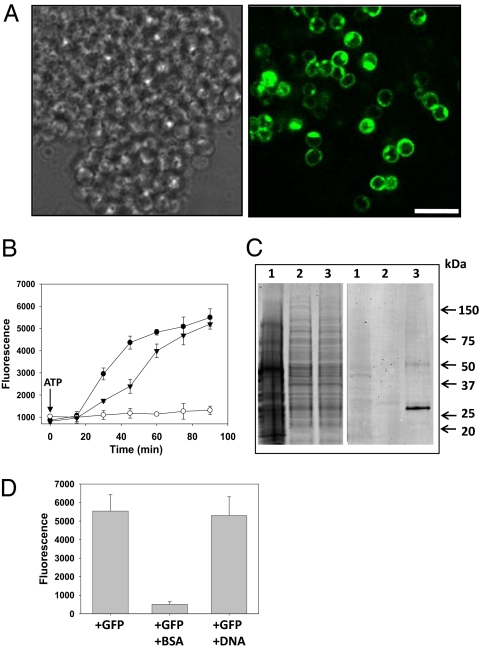
Fig. 1.
G. obscuriglobus cells uptake proteins but not DNA in an energy-dependent process. (A) Bright field microscopy and CLSM images of G. obscuriglobus cells incubated with GFP (10 μg/mL) for 60 min. Left and Right microscopy images correspond to bright field and GFP fluorescence, respectively. Because of optical sectioning effects, only some of the cells in the clump positive for GFP signal can be visualized. (Scale bar, 4 μm.) (B) Time-course experiment with GFP alone (filled circles), with GFP and 1 mM sodium azide (open circles), or with GFP and both 1 mM sodium azide and 5 mM ATP (filled triangles). After washing of G. obscuriglobus cells, GFP fluorescence was quantitatively analyzed by microplate reader assay. Error bars represent the SD from three experimental replicates. (C) Western blot analysis showing uptake of full-length GFP (≈27 kDa) by G. obscuriglobus. Cells were incubated with GFP for 1 h. After washing, cell lysates were electrophoretically separated on an SDS gel (Left), then blotted and probed with an anti-GFP antibody (Right). Lane 1, E. coli DH5α incubated with GFP; lane 2, G. obscuriglobus incubated without GFP; lane 3, G. obscuriglobus incubated with GFP. (D) Competition uptake experiment with GFP, together with either BSA or DNA. G. obscuriglobus cells were incubated for 1 h with 10 μg/mL of GFP alone, with GFP and BSA (100 μg/mL), or with GFP and plasmid DNA (100 μg/mL). Cells were then washed and analyzed for fluorescence emission by microplate reader assay. Error bars represent the SD from three experimental replicates.