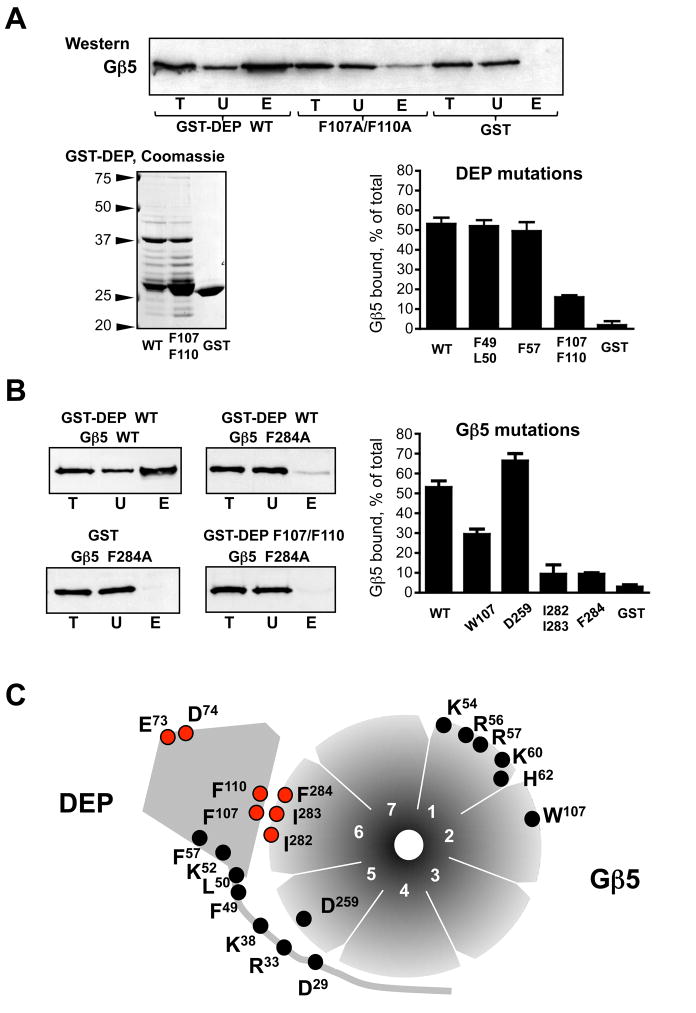
Figure 2. Mutational analysis of the DEP:Gβ5 interface.
Indicated amino acids of the RGS7 DEP domain or Gβ5 were substituted. Residues D29, R33, K38, K52, E73 and D74 were changed to the corresponding RGS9 residues, and all other residues were substituted by alanines, as described in Materials and Methods. (A) Analysis of the RGS7 DEP domain mutants. The DEP domain mutants were expressed as GST fusion proteins and analyzed by SDS-PAGE and Coomassie Blue staining. Along with the full-length GST-DEP fusion protein (~38 kDa) these preparations contained apparent degradation products, which were similar among all the DEP mutants. The wild-type and mutant GST-DEP proteins were immobilized on glutathione beads, so that the amounts of the fusion proteins were equal and corresponded to the amount of GST immobilized on the control beads. The complex of wild-type Gβ5 with the RGS7249-469 construct was expressed in CHO-K1 cells by transient transfection, and the cell lysate was subjected to the GST pull-down, as described in Materials and Methods. The western blot panel shows a representative experiment where the Gβ5-RGS7249-469 complex was tested for binding with the beads containing the GST fusion of wild type DEP domain (WT), the double-mutant F107A/F110A, or GST. The beads were washed and then eluted with SDS-containing sample buffer. The total CHO-K1 cell lysate (T), the unbound (U) and the eluate (E) fractions were probed with the anti-Gβ5 antibody, followed by ECL detection. The films were scanned and analyzed with Scion software. The bar graph summarizes the quantification of the data obtained from the entire series of experiments with all tested RGS7 DEP mutants. Material in the eluate (E) fractions was 5 times more concentrated relative to the unbound or total to ensure that the resulting ECL signal was within the linear range of detection. The values of the amount of Gβ5 present in the analyzed fractions was calculated accordingly and presented as the percentage of Gβ5 eluated from the beads relative to the total amount of Gβ5 in the cell lysate. Shown are the mean values with error bars representing standard deviation from 3–6 independent experiments. (B) Analysis of Gβ5 mutations. The indicated Gβ5 mutants were expressed in CHO cells in a complex with the RGS7249-469 construct and tested for their ability to bind to wild-type GST-DEP using the pull-down described in (A). Representative western blots show the total lysate (T) unbound (U) and eluted (E) fractions analyzed by immunoblot using the antibodies against Gβ5. The lower right panel shows the experiment where the Gβ5 F284A mutant was tested for binding with the F107A/F110A mutant of GST-DEP. The bar graph depicts quantification of the data from 3–4 independent experiments with four Gβ5 mutants (W107A, D259A, F284A and the I282A/I283A double-mutant) binding to wild-type GST-DEP or GST. (C) Diagram summarizing our mutational analysis of Gβ5 and RGS7. The seven blades of the Gβ5 “β-propeller” are numbered 1 through 7. The indicated mutants were tested in the GST pull-down assays described in A and B; the E73S/D74G mutation was described in our earlier study (21). The mutations resulting in a reduction of DEP-Gβ5 interaction are denoted by the red circles, those without a distinct phenotype are marked in black.