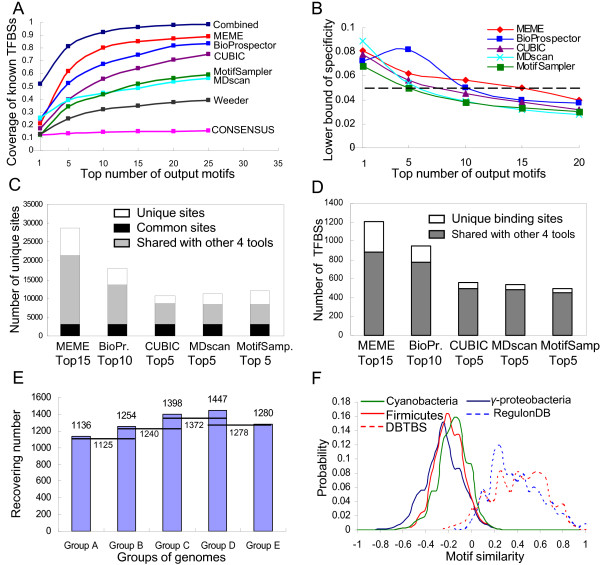
Figure 2.
Selection of parameters of the algorithm. (A). Performance of motif-finding tools returning different numbers of top predictions for recovering known TFBSs in E. coli K12 in the inter-operonic sequence sets. (B). Low bound prediction specificity of the motif-finding tools returning different numbers of top predictions for recovering known TFBSs in E. coli K12 in the inter-operonic sequence sets. (C). The number of total sites that are uniquely predicted by a tool or jointly predicted by the tool and other four tools. (D). The number of known TFBSs in E. coli K12 that are uniquely predicted by a tool or jointly predicted by the tool and other four tools. (E). Effect of the selection of different groups of target genomes on the prediction of known TFBSs in E. coli K12. (F). The distribution of motif similarity scores among the input motifs in the target genomes and that of motif similarity scores among the sub-motifs of the known motifs in E. coli K12 (RegulonDB) and B. subtilis (DBTBS).