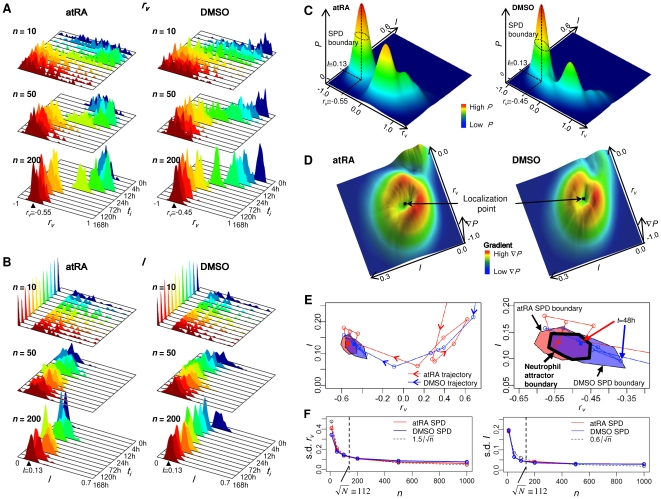
Figure 2. Determination of whole genome attractors for atRA and DMSO responses.
Temporal probability distributions of (A) rv and (B) I for atRA (upper panel) and DMSO (lower panel) for n = 10, 50, 200 genes randomly selected from whole genome. As n is increased, the distributions transit from non-localized to localized at rv≅−0.55, I≅0.13 for t≥48h (atRA) and rv≅−0.45, I≅0.13 for t≥24h (DMSO), where the bandwidth of Gaussian kernel is given by 0.02 and 0.01 for rv and I distributions, respectively. Note that rv≅−0.5 represents Pearson r>0.94 (Figure S3). Note that to visualize the localization of probability distributions at different time points, intervals are compressed into equal plot intervals. (C) 3D plot of the superposition of the probability (P) distributions (SPD) of rv and I over all time points. SPDs were estimated on discretized lattice using the MASS R library (two-dimensional kernel density estimation [34]). The boundary of SPDs for atRA (left panel) and DMSO (right panel), indicated by dotted line, is determined by selecting inflection points on the distributions, where inflection points were estimated on the lattice by selecting the points with highest gradient in 8 adjacent directions from the localization points (peak values of SPDs). Joining these points formed inflection curves for atRA and DMSO responses. (D) 3D plot of the gradient, 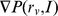 , of SPD of rv and I for atRA (left panel) and DMSO (right panel). To obtain the average SPD boundaries (inflection curves), we repeated this process 30 times. Note that I was scaled (five folds) to match gradients with rv. (E) Whole genome trajectories of rv and I for atRA and DMSO are represented by taking the average of 100 trajectories of 200 (nt) randomly chosen genes from whole genomes for t = 0, 2, 4, 8, 12, 18, 24, 48, 72, 96, 120, 144, 168h. Filled polygons indicate the SPD boundaries (inflection curve) for atRA (red) and DMSO (blue). Overlapping of the two SPD boundaries, in purple, indicates the neutrophil attractor. The lines with arrows indicate the whole genome trajectories, red for atRA and blue for DMSO. Dotted curve
, of SPD of rv and I for atRA (left panel) and DMSO (right panel). To obtain the average SPD boundaries (inflection curves), we repeated this process 30 times. Note that I was scaled (five folds) to match gradients with rv. (E) Whole genome trajectories of rv and I for atRA and DMSO are represented by taking the average of 100 trajectories of 200 (nt) randomly chosen genes from whole genomes for t = 0, 2, 4, 8, 12, 18, 24, 48, 72, 96, 120, 144, 168h. Filled polygons indicate the SPD boundaries (inflection curve) for atRA (red) and DMSO (blue). Overlapping of the two SPD boundaries, in purple, indicates the neutrophil attractor. The lines with arrows indicate the whole genome trajectories, red for atRA and blue for DMSO. Dotted curve 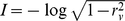 represents the linear correlation of mutual information, I, estimated by Gaussian distributions [32]–[33]. Bottom panel: enlargement of the attractor region. The thick line indicates the neutrophil attractor boundary. (F) Standard deviation of the SPDs of rv (left panel) and I (right panel) at the attractor for atRA (red) and DMSO (blue) decreases as n is increased, following the
represents the linear correlation of mutual information, I, estimated by Gaussian distributions [32]–[33]. Bottom panel: enlargement of the attractor region. The thick line indicates the neutrophil attractor boundary. (F) Standard deviation of the SPDs of rv (left panel) and I (right panel) at the attractor for atRA (red) and DMSO (blue) decreases as n is increased, following the 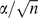 law where α = 1.5 for rv and α = 0.6 for I.
law where α = 1.5 for rv and α = 0.6 for I.