Figure 7.
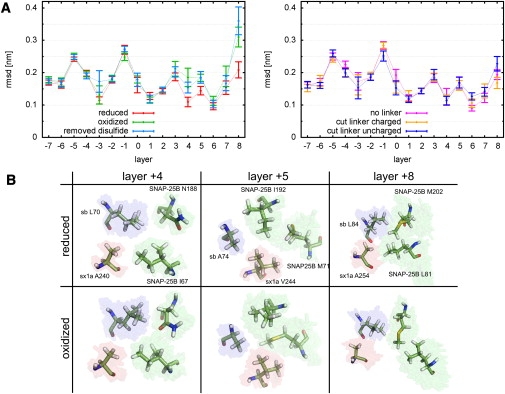
Structural deviations of the residues which contribute to the hydrophobic layers for the six simulations discussed in the text. (A) Shown are the means and the standard errors for the RMSD values calculated from the final 40 ns of each simulation. The RMSD of each layer was calculated after fitting the positions of the Cα-atoms to those of the modeled structure. (B) (Stick representation) For “reduced” and “oxidized” setups SNAP-25B, syntaxin 1A (sx1a), and synaptobrevin (sb) residues contributing to layers +4, +5, and +8. (Solid sticks) One conformation at the end of a trajectory. (Colored region) An ensemble from the final 40 ns of all trajectories.
