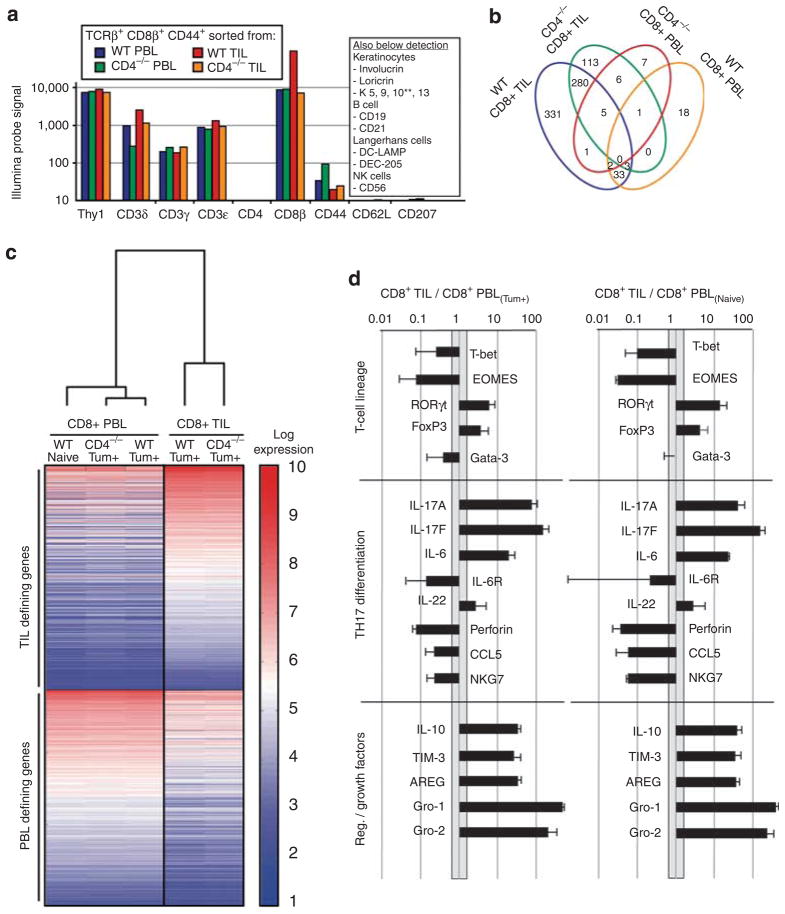
Figure 2. Illumina expression analysis of CD8+TCRβ+CD44HICD62LO TIL and PBL populations.
mRNA from highly purified (>98%) CD44HICD62LLOTCRβ+CD8+ tumor-infiltrating lymphocyte (TIL) and peripheral blood lymphocyte (PBL) from tumor-bearing (Tum+) CD4−/− and Tum+ wild-type (wt) mice were analyzed by Illumina Sentrix bead chips (~47,000 genes). (a) Absolute probe hybridization values were consistent with high-fidelity purifications. (b) Relative to the naive PBL, a total of 800 (~1.7%) genes were found to be >10-fold differentially expressed by the TIL and PBL sorts in Tum+ mice. The vast majority of these (91%) were within the TIL sets. (c) Hierarchical clustering analysis showed that the PBL populations isolated from tumor-bearing (Tum+) mice were similar to phenotypically equivalent (CD44HICD62LLOTCRβ+CD8+) effector T cells purified from PBL of naive animals. The dendrogram shows that the primary clusters based on global gene expression differentiate between PBL and TIL, whereas the heatmap (red, high expression; blue, low expression) highlights some of the most differentially expressed genes defining these two major groups. (d) Analysis of the most differentially expressed genes unique to CD8+ T-pro (based on absolute probe values) showed differentiation along the RORγt pathway, poor cytotoxic potential, and production of regulatory mediators/growth factors.