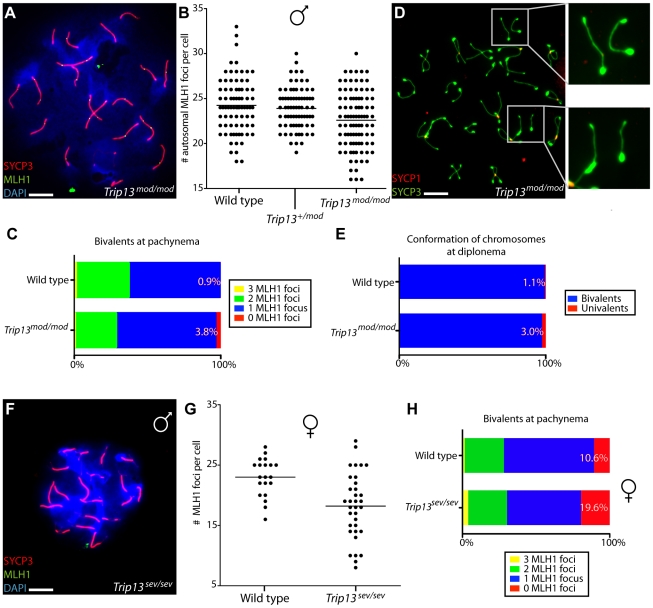
Figure 8. Reduced COs and chiasmata in Trip13-deficient spermatocytes and oocytes.
(A) Spread chromosomes from a Trip13mod/mod pachytene spermatocyte stained for DNA (DAPI), SYCP3, and MLH1 to mark the positions of COs. Bar = 10 µm. (B) Quantification of the number of autosomal MLH1 foci per pachytene cell. Means are shown as horizontal lines. Results were as follows (mean ± sd): 24.2±3.1 (wild type, six mice); 23.9±2.3 (Trip13+/mod, seven mice); 22.6±3.2 (Trip13mod/mod, four mice). (C) Percentages of bivalents in wild-type and Trip13mod/mod pachytene spermatocytes with the indicated numbers of MLH1 foci. (D) Spread chromosomes from a Trip13mod/mod diplotene spermatocyte stained for SYCP3 and SYCP1. Note the presence of achiasmate bivalents, some of which are presented in the zoomed images on the right. Bar = 10 µm. (E) Percentages of achiasmate chromosome pairs at diplonema in wild-type and Trip13mod/mod spermatocytes. (F) Absence of MLH1 foci in a pachytene-like Trip13sev/sev spermatocyte. Bar = 10 µm. (G) Quantification of MLH1 focus numbers in pachytene or pachytene-like oocytes. Means are indicated by the horizontal line. Results were as follows (mean ± sd): 23.0±3.2 (wild type); 18.2±5.5 (Trip13sev/sev). (H) Percentages of bivalents with the indicated numbers of MLH1 foci. This analysis provides a conservative estimate of the defect in Trip13sev/sev oocytes, because only cells with ≥20 total foci were considered in order to minimize possible secondary effects of differences in the timing of progression through meiosis.