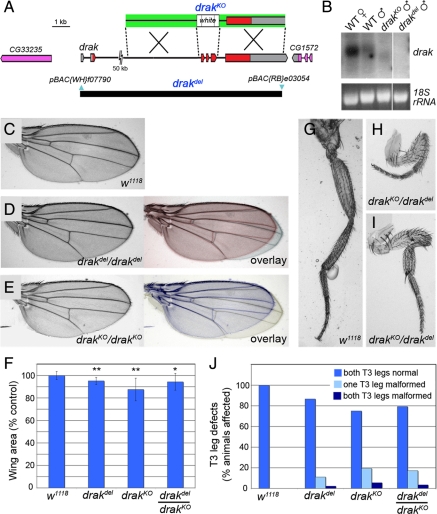
Figure 2.
drak mutants have a “malformed' phenotype. (A) Map of the drak locus and defined deletions. The relative positions of noncoding (gray) and coding (red) exons of drak and the upstream (CG33235) and downstream (CG1572) neighboring genes are indicated. A schematic of the targeting construct used to generate the drakKO allele is shown above (arms of homology, green). The positions of the piggyBac insertions (blue) used to make the drakdel allele and extent of the resulting deletion (black bar) are shown below. (B) Northern blot analysis of RNA extracted from w1118 female and male adult flies (WT), and drakKO and drakdel mutant adult males. 18S rRNA was stained to ensure equal loading. Images are all from the same exposure of a single blot/gel with intervening lanes removed. (C–E) Wings from wild-type (C), drakdel/drakdel (D), and drakKO/drakKO (E) mutant flies. Overlays of drak mutant and wild-type wings are in color. (F) Areas of wild-type and drak mutant wings (average ± SD), based on measurement of 10 wings for each genotype. t test versus wild-type: *, p <0.05; **, p <0.001. (G–I) T3 legs from wild-type (G) and drakKO/drakdel (H and I) flies. (J) Quantification of frequency of malformed T3 leg phenotype in wild-type (n + 30), drakdel/drakdel (n + 45), drakKO/drakKO (n + 36), and drakdel/drakKO (n + 29) mutant flies.