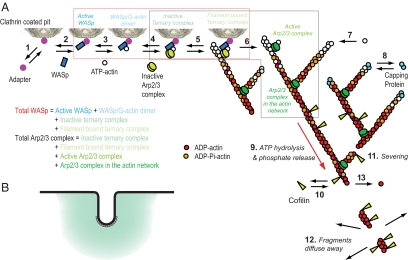
Figure 1.
Diagram illustrating the reactions assumed in the mathematical model and simulations of actin filament assembly and disassembly at sites of endocytosis. (A) Numbered reactions in the model: 1, adapter binds clathrin; 2, Wsp1p binds adapter; 3, actin binds Wsp1p; 4, Arp2/3 complex binds Wsp1p (ternary complex); 5, inactive ternary complex binds filament; 6, filament activates ternary complex to initiate a branch; 7, ATP-actin elongates filament barbed ends; 8, capping protein blocks barbed ends; 9, actin hydrolyzes bound ATP and dissociates phosphate; 10, cofilin binds ADP-actin filaments; 11, filaments sever; 12, filament fragments diffuse away; and 13, ADP-actin subunits dissociate from free pointed ends. The red box encloses all WASp species in the model (Active WASp, dark blue; WASp/G-actin monomer, light blue; Inactive ternary complex, turquoise; Filament bound ternary complex, light green) for comparison with the amount of Wsp1p measured experimentally. The black box encloses all Arp2/3 complex species in the model (Inactive ternary complex, turquoise; Filament bound ternary complex, light green; Active Arp2/3 complex, green; Arp2/3 complex in the actin network, dark green) for comparison with ARPC5 measured experimentally. (B) A schematic diagram of actin network assembly at the site of endocytosis. Black line represents cell membrane, gray boxes represent clathrin at the tip of invagination, and teal represents actin network in a 300-nm sphere around endocytic invagination. The intensity of teal shading represents postulated gradient of density of actin network.