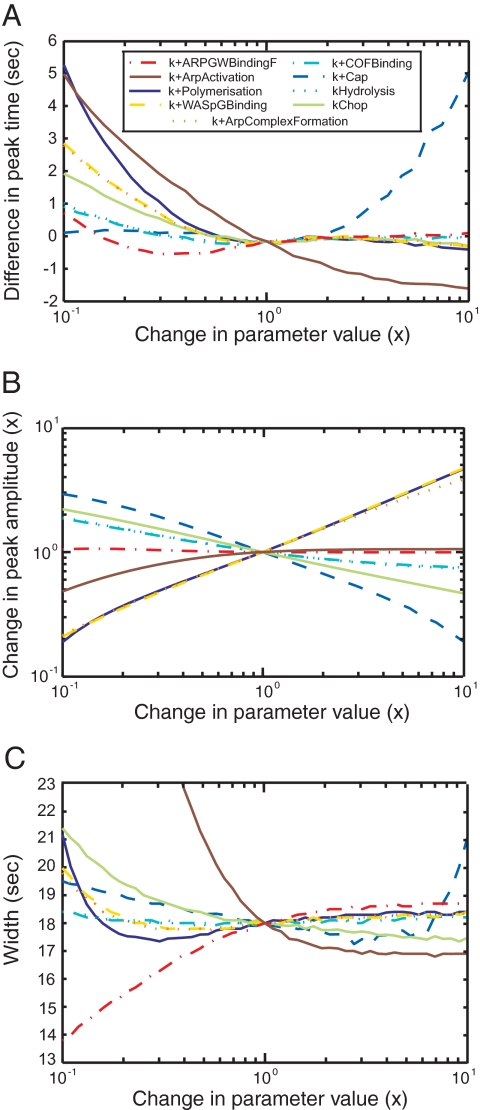
Figure 5.
Sensitivity analysis. The model was simulated with each parameter varied ± 1 order of magnitude to determine the sensitivity of the time course of actin accumulation and dispersal on parameter values. (A) Times of protein peaks. (B) Amplitudes of protein peaks. (C) Widths of protein peaks, defined as the difference between the times when the signal reaches 25% of its peak value during the assembly and during the disassembly. Color code for varied parameters: yellow dashed line, kWASpGBinding+; orange dotted line, kArpComplexFormation+; red dotted and dashed line, kARPGWBindingF+; brown plain line, kArpActivation+; dark blue plain line, kPolymerization+; blue dashed line, kCap+; light blue dotted line, kHydrolysis; light blue dotted and dashed line, kCOFBinding+; plain light green line, kChop. Parameters for the reverse reactions are not shown, because varying these parameters has little effect on the time courses of proteins in patches.