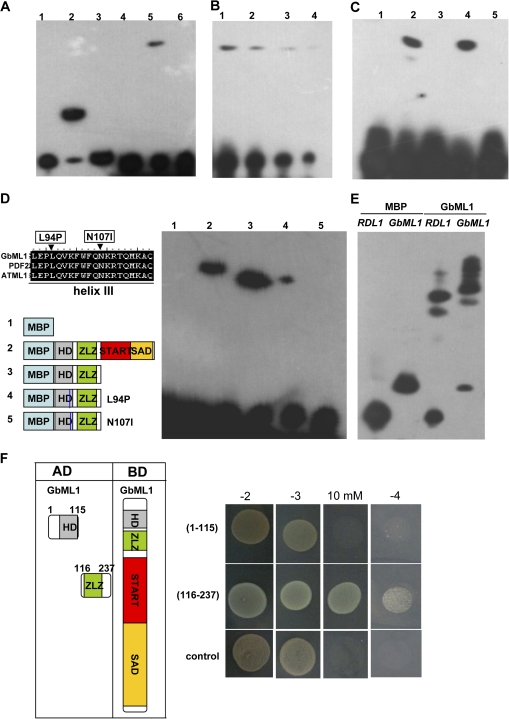
Fig. 2.
GbML1 binds to the L1 box in vitro. (A) Interaction of GbML1 protein with the L1 box. 1–3: Controls provided by the kit. 1, No protein, biotin-ENBL probe; 2, ENBL, biotin-ENBL probe; 3, ENBL, biotin-ENBL probe and 100× ENBL cold competitor. 4–6, binding assay for GbML1;. 4, MBP, biotin-L1 probe (L1: TGTAAATGCACCTGCAACACA); 5, GbML1, biotin-L1 probe; 6, GbML1, biotin ml1 probe (mL1:TGTAAGGGCACCTGCAACACA). (B) Reduced probe concentration results in weaker binding. 2, 2×dilution; 3, 4×dilution; 4, 8× dilution. (C) HD-ZLZ domains are required and sufficient for binding to the L1 box. Binding reaction includes biotin-labelled L1 box probe with different MBP fusion proteins: 1, MBP protein; 2, MBP-GbML1 protein; 3, MBP-HD protein; 4, MBP-HD-ZLZ protein; 5, MBP-START–SAD protein. (D) Point mutations in the third helix affect the binding of HD-ZLZ to L1 box. Left: Diagram of the point mutation and proteins used in EMSA. Right, EMSA assay. The number above is correlated to the number of the proteins diagramed. (E) GbML1 binds to the L1 box containing promoters from cotton. (F) ZLZ domain confers homodimer formation. left: Diagram of different constructs used for the yeast two-hybrid assay; right, yeasts harbouring BD-GbML1/AD-HD or BD-GbML1/AD-ZLZ grown on selective plates as indicated. Control medium: –2 (SD/-T-L); selective medium: –3 (SD/-T-L-H), 10 mM (–3 supplemented with 10 mM 3-AT), –4 (SD/-T-L-H-A). The control is yeast transformed with BD-GbML1/AD.