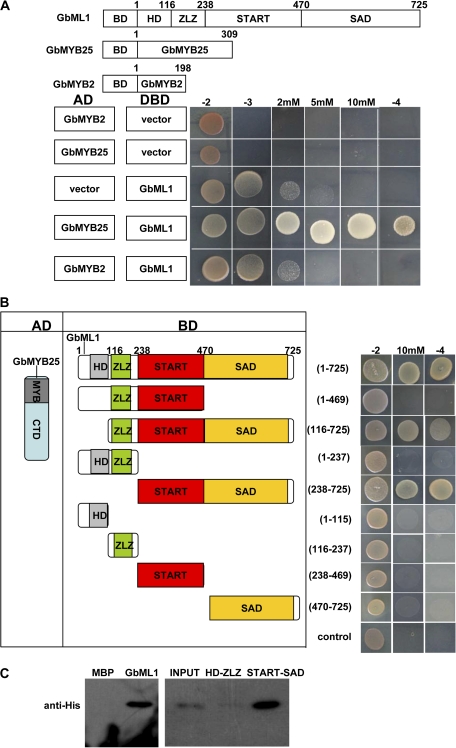
Fig. 4.
START–SAD domain of GbML1 are required and sufficient for the interaction with GbMYB25 protein. (A) GbML1 could bind to GbMYB25 but not to GbMYB2. (Top) diagram of different constructs used in this panel. (Bottom) Y2H assay. Yeast cells harbouring the constructs shown on the left were spotted on control medium (–2) and selective medium (–3; –3 with 2 mM 3-AT, 5 mM 3-AT, or 10 mM 3-AT; –4). (B) Mapping of domains of GbML1 to bind GbMYB25. As shown on the left, different truncated versions of GbML1 protein were fused with the BD domain and co-transformed into yeast with AD-GbMYB25. The yeast cells harbouring the bait and prey were spotted on the selective medium. As shown, both the START domain and the SAD domain are required for the interaction with GbMYB25. Yeast cells habouring BD and AD-GbMYB25 constructs were used as the negative control. (C) In vitro pull-down assay of MBP, full-length or truncated GbML1 proteins with HIS-GbMYB25 fusion protein. The full-length or truncated MBP-GbML1 fusion proteins are used as baits to pull down the His-GbMYB25 fusion protein from the induced cell extracts. MBP protein is assayed as a negative control. Immunoblot detection of prey protein is with His antibody. Arrow indicates the corresponding target proteins.