Abstract
Microcephaly, mental retardation and congenital retinal folds along with other systemic features have previously been reported as a separate clinical entity. The sporadic nature of the syndrome and lack of clear inheritance patterns pointed to a genetic heterogeneity. Here, we report a genetic analysis of a female patient with microcephaly, congenital bilateral falciform retinal folds, nystagmus, and mental retardation. Karyotyping revealed a de novo pericentric inversion in chromosome 6 with breakpoints in 6p12.1 and 6q21. Fluorescence in situ hybridization analysis narrowed down the region around the breakpoints, and the breakpoint at 6q21 was found to disrupt the CDK19 gene. CDK19 was found to be expressed in a diverse range of tissues including fetal eye and fetal brain. Quantitative PCR of the CDK19 transcript from Epstein–Barr virus-transformed lymphoblastoid cell lines of the patient revealed ~50% reduction in the transcript (p = 0.02), suggesting haploinsufficiency of the gene. cdk8, the closest orthologue of human CDK19 in Drosophila has been shown to play a major role in eye development. Conditional knock-down of Drosophila cdk8 in multiple dendrite (md) neurons resulted in 35% reduced dendritic branching and altered morphology of the dendritic arbour, which appeared to be due in part to a loss of small higher order branches. In addition, Cdk8 mutant md neurons showed diminished dendritic fields revealing an important role of the CDK19 orthologue in the developing nervous system of Drosophila. This is the first time the CDK19 gene, a component of the mediator co-activator complex, has been linked to a human disease.
Introduction
Congenital retinal fold (ablatio falciformis congenita) refers to a congenital malformation that is characterized by the presence of a raised retinal fold extending radially from the posterior pole to the fundus periphery. It is reported that congenital retinal fold is not a single clinical entity, but can be a manifestation of a diverse group of conditions (van Nouhuys 1982a). Familial exudative vitreoretinopathy (FEVR; MIM 133780) and microcephaly are frequently associated with congenital retinal fold (van Nouhuys 1981, 1982a). In OMIM two separate entries (MIM, 152950 and 156590) describe several reports in which the patients suffer from microcephaly, mild to moderate mental retardation, chorioretinopathy or congenital retinal fold, and lymphedema. The reports depict a heterogeneous clinical spectrum of these features with an uncertain inheritance pattern pointing towards a heterogeneous genetic etiology for this rare developmental syndrome.
In four eyes of three patients from two large families with FEVR, congenital retinal folds were found (van Nouhuys 1982a). In the same report, it was mentioned that the formation of congenital retinal folds is the consequence of a developmental disorder of retinal vasculature during the last few months of intrauterine life and the folds may even develop after birth. The clinical picture and familial occurrence of many cases of congenital retinal folds previously described were suggestive of FEVR. Nishimura et al. (1983) likewise found falciform retinal folds as a sign of FEVR.
Several reports on patients with bilateral congenital retinal folds mention the presence of microcephaly with varied degree of mental retardation. The majority of the patients showed bilateral falciform folds extending from the optic disc to the inferior-temporal quadrant of the fundus periphery. The similarity of the retinal folds in this group of microcephalic patients suggested an independent clinical entity (van Nouhuys 1982b).
Gartner (1941) and Masuda (1962) reported two independent exudative vitreoretinopathy patients, of which the parents were consanguineous, pointing to autosomal recessive transmission. On the other hand, Pfluger (1885) and Jarmas et al. (1981) reported patients of which one of the parents shared some, but not all of the clinical features, suggesting autosomal dominant transmission with variable expression. Recently, Nguyen et al. (2005) reported on a boy with congenital microcephaly, juvenile retinal dystrophy but without mental retardation. His eye examination revealed an optically empty vitreous and poorly developed fovea, pointing to the etiological relationship between this condition and FEVR.
In this study, we report the clinical and molecular findings in a female patient with bilateral congenital retinal folds and microcephaly with mild mental retardation. By cytogenetic and molecular genetic analyses, we have found that one copy of the CDK19 gene, located at 6q21, is disrupted in the patient as a result of a de novo pericentric inversion. In addition, we have used Drosophila as a model system and identified a role of this gene in the developing nervous system.
Materials and methods
Clinical investigation
An informed consent was obtained from our index patient as well as from other individuals who has been recruited for the present study. Ophthalmological examination of the index patient has been performed on several occasions from 1979 to 2008. The following methods have been used: determination of visual acuity (Landolt C and Cardiff tests) and contrast sensitivity, applanation tonometry (Haag-Streit, Koeniz, Switzerland), gonioscopy, slit lamp examination of the anterior segment and vitreous (Rodenstock slit lamp, Munich, Germany), indirect ophthalmoscopy (Zeiss ophthalmoscope, Oberkochen, Germany), biomicroscopy (Rodenstock slit lamp), and digital fluorescein angiography of the fundi (Zeiss FF450 fundus camera, Win station software, Medivision-OIS, Sacramento, USA) and Goldmann perimetry (Haag-Streit perimeter, Munich, Germany). The patient has also been subjected to a full pediatric, neurological and otolaryngological examination.
Cell culture
Human B-lymphocytes were immortalized by transformation with the Epstein–Barr virus (EBV) according to established procedures (Wall et al. 1995). EBV-transformed lymphoblastoid cell lines (EBV–LCLs) of the patients and controls were grown to a density of 0.7 million cells per ml RPMI 1640 medium (Gibco, Breda, The Netherlands) containing 10% (v/v) fetal calf serum (Sigma, The Netherlands), 1% penicillin–streptomycin (Gibco, The Netherlands), and 1% GlutaMAX™ (Gibco, The Netherlands). Twenty-five million cells were harvested by centrifugation at 200×g for 5 min at room temperature and re-suspended in 500 μl 8 mM Na2HPO4, 2 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl pH 7.2 (PBS).
Fluorescence in situ hybridization (FISH)
Non-radioactive in situ hybridization on human metaphase spreads was performed essentially as described earlier (de Bruijn et al. 2001). The probes to be tested by FISH were selected from an array of 32,000 BAC probes covering the human genome (32 K BAC array). The analysis was performed on methanol/acetic acid fixed cells of cell lines from individuals under study. In short, BACs are biotin labeled by nick translation and then purified, and ethanol precipitated together with a 30-fold excess of Cot-1 DNA (Life Technologies). Immunocytochemical detection of the hybridizing probe was achieved using fluorescein isothiocyanate (FITC)-conjugated avidin followed by one application step of goat-anti-avidin conjugated with FITC.
The commercially available (Vysis) reference probe for centromere of chromosome 6, gives an orange fluorescence signal. A Zeiss Axiophot-2 microscope equipped with the appropriate fluorescence filters was used for analysis of the fluorescence signals. Images were captured by a Leica DC 350 FX camera using Leica CW 4000 software package.
Semi-quantitative RT–PCR and real-time quantitative RT–PCR analysis
Total RNA was isolated from EBV-LCLs using the RNeasy midi-kit (Qiagen, Venlo, The Netherlands) according to the manufacturer’s protocol. The quality and quantity of the RNA was determined by spectrophotometric measurement and gel electrophoresis. cDNA was synthesized from 2.0 μg total RNA using random hexamers (Amersham, The Netherlands), and M-MLV reverse transcriptase (Invitrogen, The Netherlands) in a total volume of 85.0 μl. After the synthesis, the cDNA was purified using QIAquick PCR purification columns (Qiagen, Venlo, The Netherlands) and eluted in 30 μl milliQ water.
To determine the expression profile semi-quantitative RT–PCR was performed using primer pairs that amplify parts of CDK19 and ELOVL5 transcripts. The primer sequences for CDK19 was 5′-CGGAACCTATTTTTCACTGTCG-3′ (exon 7) and 5′-TGTGGGATATTCTGGCATCTT-3′ (exon 8) and that for ELOVL5 was 5′-CATTCAGACCTACAACAAGAAAGG-3′ (exons 6–7) and 5′-GGCTTCACATTGTTTTCCAGGG-3′ (exon 7). The PCR was performed for 30 cycles using RNA (cDNA) from 12 different human tissues. GAPDH was used as a reference gene.
For real-time quantitative RT–PCR (QPCR), 40× dilutions of the cDNA were made and 5.0 μl from the diluted stock was used for each PCR reaction. Primers were designed using Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Amplicon lengths were typically kept between 80 and 130 bp. Quantitation was performed using SYBR-Green-based relative quantification strategies according to the manufacturer’s protocol (Bio-Rad, The Netherlands). PCR reactions were performed on an automatic iCycler (MyiQ single color Real-Time detection System, Bio-Rad, The Netherlands). Initially, the primer conditions were optimized and a standard curve was determined for six different dilution points (10×–320×) until for each of the primers PCR efficiencies close to 100% were obtained. For calculation purpose, all the PCR efficiencies were assumed to be exactly 100%. Two independent cDNA syntheses were made from each RNA sample and duplicate QPCR reactions were performed for each cDNA to ensure reproducibility. The following condition was used for all the QPCR experiments: initial denaturation for 3 min at 95.0°C, followed by 15 s at 95.0°C and 30 s at 60.0°C for 40 cycles (QPCR data collection). The products were then denatured at 95.0°C for 1 min and cooled to 65.0°C and kept for 1 min (melt curve data collection).
Ten different EBV–LCLs from 10 unrelated control samples from the Netherlands were used to determine the normal range of expression for CDK19 and ELOVL5. The mean expression level was used for normalization. A recent report has systematically identified stable gene expressions in EBV–LCLs (de Brouwer et al. 2006) for use as reference genes for normalization purpose. We have selected the most stable genes from this study, and after using Genorm (Vandesompele et al. 2002) GUSB and RNF111 were found to be two most stable genes in our experiments. The geometric mean of the expression of GusB and RNF111 was used as the value for reference genes for normalization.
The differences in expression between patients and controls were calculated using the 2ΔΔCt method (Livak and Schmittgen 2001; Pfaffl 2001). By definition, C t values equal the required number of amplification cycles to reach the threshold fluorescence level (automatically determined); ΔC t implies the difference in the C t values between the reference gene (mean) and the gene of interest (CDK19 or ELOVL5) and ΔΔC t represents the difference between two ΔC t values obtained from patient and the control (mean). For statistical analysis of the QPCR data, the p values were calculated in the p value calculator (www.graphpad.com/quickcalcs/PValue1.cfm) available in the public domain.
Drosophila studies
The UAS-Gal4 system (Brand and Perrimon 1993) was used for induced expression of dsRNA and for labeling of neurons with GFP. The UAS-Cdk8 RNAi lines were obtained from the National Institute of Genetics Fly Stock Center (stock numbers 10572R-1 and 10572R-3, Kyoto, Japan). These stocks contain an inducible 500 base pair dsRNA homologous to the Cdk8 coding region (exon 1 and part of exon 2). Details on the sequence and the absence of predictable off-targets are available at http://www.shigen.nig.ac.jp/fly/nigfly/index.jsp. The ubiquitous promoter-Gal4 line y 1 w * ;P{Act5C-GAL4}25FO1/CyO,y +(Act-Gal4) and y 1 w*;P[GawB]477,P[UAS-mCD8::GFP.L]LL5 (477-Gal4, UAS-mCD8::GFP), which expresses a membrane targeted mCD8-GFP fusion protein in Drosophila type 4 multiple dendrite (md) neurons (Grueber et al. 2003), were obtained from the Bloomington Drosophila stock center at Indiana University (stock numbers 4414 and 8746, Bloomington, USA).
For analysis of Cdk8 knockdown efficiency Act-Gal4 was balanced over a CyO-GFP chromosome. Act-Gal4/CyO-GFP flies were then crossed to wild-type flies and to flies harboring the homozygously viable UAS-Cdk8 RNAi insertion. Third instar larval progeny that received the Act-Gal4 transgene were identified based on the lack of GFP staining. Drosophila total RNA was extracted in triplicate from third instar larvae of the genotypes UAS-Cdk8 RNAi /+; Act-Gal4/+ and Act-Gal4/+ using the RNA Lipid Tissue mini kit (QIAGEN, Venlo, The Netherlands). RNA was transcribed into cDNA using the iScript cDNA Synthesis Kit (Bio-Rad, Veenendaal, The Netherlands) and cDNA was purified using Nucleospin Extract II PCR clean-up kit (Macherey–Nagel, Düren, Germany). SYBR-Green-based real-time quantitative PCR analysis was performed on a 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using Power SYBR-Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. Relative expression levels were calculated as described above using the Act5c gene as a reference.
For analysis of dendritic development 477-Gal4, UAS-mCD8-GFP/CyO flies were crossed to wild type and UAS-Cdk8 RNAi flies. Late third instar larvae expressing mCD8-GFP were dissected and immunostained using a rat anti-mCD8 antibody (1/200) (Invitrogen, Breda, The Netherlands) and an anti-rat Alexa Fluor 488 conjugated secondary antibody (1/1000) (Invitrogen, Breda, The Netherlands). A Leica DM-IRE2 confocal microscope (Leica Microsystems, Rijswijk, The Netherlands) was used to create images from a region of 375 × 375 μm between the denticle belts of the abdominal segments 3–5 of the wandering third instar larvae. Each larval hemi-segment contains four clusters of md sensory neurons (ventral, ventral’, lateral and dorsal), which are morphologically characterized and are positioned in a highly stereotypical pattern (Grueber et al. 2002). In this study, we have analyzed the morphology of the solitary type 4 md dendritic arborization neuron in the ventral cluster called vdaB (ventral dendritic arborization neuron B). These neurons were chosen for analysis because of their highly stereotyped dendritic arbour that is amenable to quantitative morphological analysis. Microscopic images were captured using a 40× lens. The field of view was always confined by the denticle belts, on the upper (anterior) and lower (posterior) margin of the image, with the cell body of the vdaB neuron positioned approximately in the middle of the field of view from left to right (Fig. 5d). Dendrite ends were counted manually from stacked images for the entire field of view. Ten images were quantified for each genotype and the mean number of dendrite ends was compared using a one sample t test.
Fig. 5.
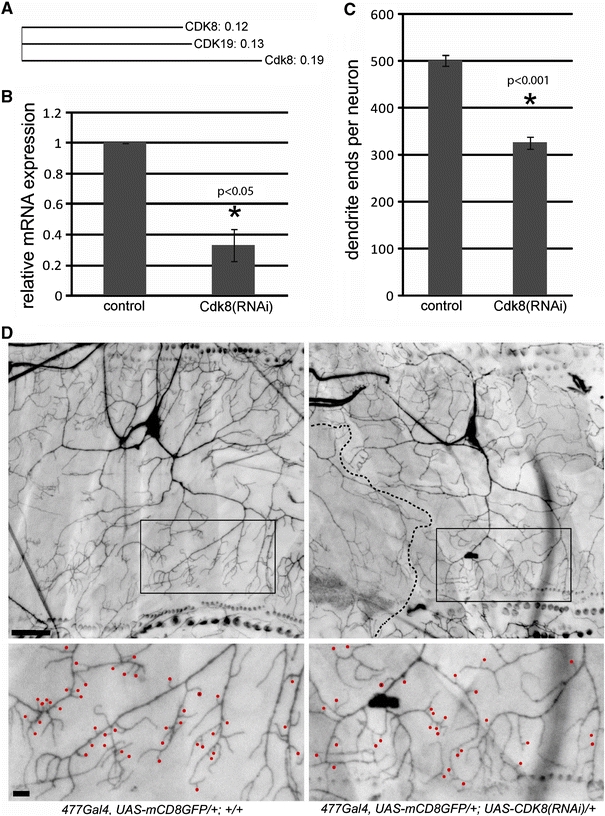
Functional analysis of the Drosophila CDK19 ortholog, Cdk8. a An unrooted phylogenetic tree comparing CDK8 protein family members in human (CDK8 and CDK19) and Drosophila (Cdk8) by the neighbor-joining method. b Relative expression of Cdk8 mRNA was measured by quantitative PCR. Cdk8 mRNA is reduced by 68% in Cdk8(RNAi) flies (genotype ActGal4/+; UAS-Cdk8(RNAi)/+) when compared with controls (genotype ActGal4/+; +/+). c Dendrite ends were counted manually from confocal images of type 4 md neurons (d). Dendritic branching was reduced by 35% in Cdk8(RNAi) flies (genotype 477-Gal4,UAS-mCD8::GFP/+; UAS-Cdk8(RNAi)/+) when compared with controls (genotype 477-Gal4,UAS-mCD8::GFP/+; +/+). d Representative images of ventral type 4 md neurons located in the larval skin. Cdk8 knockdown in these cells (right panel) causes a decrease in dendritic branching (quantified in c) and restriction of the overall dendritic arbor when compared with controls (left panel). The stippled line shows an area of restricted growth where a neighboring neuron (the v’ada type 4 md neuron from the ventral’ cluster) has grown. This altered neuronal morphology is not observed in controls. Red dots in the bottom panel show dendritic protrusions of <10 μm in length. These are more common in control neurons when compared with Cdk8-knockdown neurons (44 as compared to 31 in this one representative panel). Posterior is up. Scale bar 50 μm in the upper panel and 10 μm in the lower panel
Results
A female patient with congenital retinal fold and microcephaly
The birth weight of the girl had been 2,350 g and the neonate was not treated with oxygen, a risk factor for retinopathy of prematurity. The pregnancy had been of normal duration, but had been complicated by an ovarian cyst, which had to be removed surgically soon after parturition. Her cranial circumference at birth was only 29 cm and remained far below the P10 curve on several subsequent occasions. Café-au-lait like pigmentations of the skin were found scattered over her body. Other changes that might be consistent with neurofibromatosis were not found. From infancy, a moderate psychomotor retardation and some hearing loss was found. An adenotomy and tonsillectomy were performed at age 4 after some recurrent upper airway infections. About 20 dB of hearing loss was diagnosed due to perceptive dysfunction, which did not show progression. MRI scans performed when she was an adult did not show congenital or acquired cerebral anomalies. The patient had been followed at regular intervals (once a year) for the last 29 years without any significant progression of her visual and psychomotor conditions. At the age of 1 year, she had been referred to an ophthalmologist for a convergent position of the left eye. She was found to have horizontal nystagmus when she was 2 years old. Fundus examination showed bilateral falciform retinal folds running from the optic nerve head to the temporal periphery. Both folds were attached by white fibrous tissue to the pars plana and temporal equator of the lens (Fig. 1).
Fig. 1.
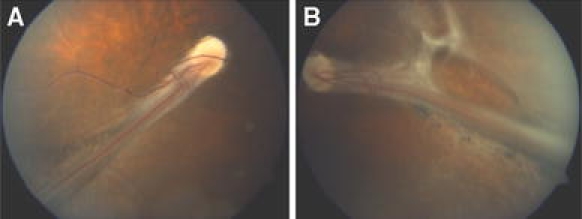
Fundus abnormalities in both eyes of the patient. a Right eye: posterior pole of fundus. A falciform retinal fold extends from the optic disc to the inferior-temporal quadrant of the fundus. Note the absence of the macula and abnormal course of retinal vessels. b Left eye posterior pole of the fundus. A falciform retinal fold extends from the disc in temporal direction. The retina superior to the fold shows some fibrous tissue
At the age of 14 years, visual acuity of the right eye and left eye was limited to finger counting. The left eye showed a convergent position. Both eyes showed a marked horizontal nystagmus. Cyclopic refraction showed a moderate hyperopic astigmatism of both eyes, but was difficult to determine because of poor fixation due to the prominent folds in the posterior poles. The anterior segments of both eyes showed a clear cornea with normal dimensions. No abnormalities of the iris and pupils were noted. Both lenses showed some capsular and sub-capsular opacity on the temporal side. The vitreous body of both eyes showed syneresis with some optically empty lacunae. Some preretinal membranes were noted temporally in both eyes. The right fundus showed marked deformation of the retinal vascular network in the posterior pole. A prominent retinal fold extended from the optic disc in inferior-temporal direction to the fundus periphery where it ended in a local mass of white fibrous tissue attached to the pars plana (Fig. 1a). The macula was not visible and several retinal vessels extended from the disc in the retinal fold. In the posterior retina, a number of retinal vessels were visible outside the fold. Some of the vessels were seen to end in terminal ramifications. The peripheral retina presented an atrophic appearance and seemed avascular. No demarcation line could be observed between vascular and avascular retinal areas. Remnants of hyaloid vasculature could not be observed. The left fundus showed a similar falciform retinal fold running from the optic nerve head towards the temporal fundus periphery, where it disappeared in a white tissue mass attached to the lens equator and the pars plana (Fig. 1b). The retina showed several pigmentations on either side of the fold and some fibrous epiretinal tissue especially superior to the fold.
At 38 years, gonioscopy was repeated because of some flattening of the anterior chamber of the left eye and moderate narrowing of the angle was noted. To prevent angle closure, an iridectomy was done with an Nd-YAG laser. No elevated intraocular pressure has been noticed before or after this procedure. Until her current age visual functions seemed to be stable. Eye examination including fundus biomicroscopy in the mother and father of the patient did not reveal significant abnormalities. A few maternal relatives showed unilateral visual impairment due to high myopia.
The patient was also found to have night blindness and lymphedema. She was subjected to several pediatric and neurological examinations at different ages. At the age of 41 years, type 2 diabetes was diagnosed and treated with diet and oral medication.
A de novo pericentric inversion in chromosome 6
Karyotyping revealed a pericentric inversion in chromosome 6 of the patient involving 6p11.1 and 6q21 (Fig. 2a). The patient otherwise had a normal female karyotype. The parents also showed normal karyotypes indicating that the patient did not inherit the chromosomal rearrangement.
Fig. 2.
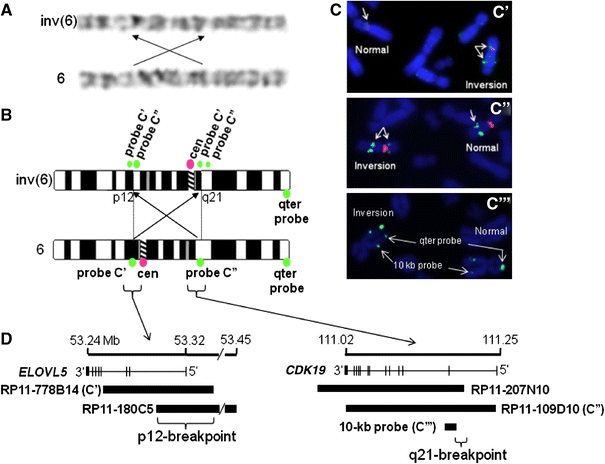
Schematic representation of breakpoints on chromosome 6. a Karyotyping showed a pericentric inversion in chromosome 6 which is schematically depicted in b below; b schemes of chromosome 6 for both the derivative (inverted) and the normal copy. The relative position and size of the FISH probes used are marked with corresponding colors on the right; c results of the FISH experiments performed with probes of the p-arm (top panel) and the q-arm (middle panel). In both cases, one of the copies (marked ‘inversion’) show a split signal positioning the breakpoint within the probe. The pink colored signal is from the centromeric probe for chromosome 6. The lower panel shows the fine mapping of 6q21 breakpoint using a PCR product as FISH probe; d schematic summary of the entire breakpoint analysis along with the genes involved in or near the breakpoints and their genomic coordinates. The cartoon shows that the inversion disrupts the CDK19 gene on 6q21, but not ELOVL5 on 6p12
To rule out other large copy number abnormalities in the patient’s genome and to narrow down the inversion breakpoints in the patient, comparative genomic hybridization was performed which did not show any clone to deviate significantly from the normal range of variation (data not shown). The karyotype for the derivative chromosome 6 indicated the breakpoint at 6p to be very close to the centromere. A location of the p-arm breakpoint in the centromere was excluded using FISH analysis of a centromeric probe for chromosome 6 (data not shown). Four BAC clones from 6q21, uniformly distributed across an 11-Mbp region, and six probes covering 10 Mbp at 6p11–p12 were selected from the 32K BAC array. Probes RP11-109D10 (C″ in Fig. 2) and RP11-778B14 (C′ in Fig. 2) were found to be broken at 6q21 and 6p12.1, respectively (Fig. 2c). To confirm these findings, additional probes from both arms of chromosome 6 were selected which overlapped with the clones mentioned above. These clones, RP11-180C5 (6p12.1) and RP11-207N10 (6q21) also showed a split signal on the derivative chromosome mapping the breakpoints to 220 kb regions on either side of the centromere.
Genes near the breakpoints
A more detailed inspection of the genomic regions covered by these BAC clones revealed that at 6p12.1 both RP11-778B14 and RP11-180C5 partially overlapped the ELOVL5 gene (Fig. 2d). Similarly, at 6q21, RP11-109D10 completely and RP11-207N10 partially overlap CDK19 (Fig. 2d). CDK19 and ELOVL5 were further investigated. Both CDK19 and ELOVL5 were found to have a wide range of tissue expression including fetal eye and fetal brain—the two tissues most likely to be involved in the developmental problems observed in the patient (Fig. 3).
Fig. 3.
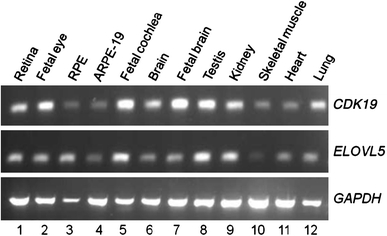
Expression profile of CDK19 and ELOVL5. The normal expression profile of CDK19 and ELOVL5 was determined in 12 human tissues. Both genes are expressed in almost all tissues. Note the higher expression of CDK19 in fetal eye (lane 2) and fetal brain (lane 7) as compared to ELOVL5. GAPDH was used as a reference gene. The PCR was done for 30 cycles
CDK19 is disrupted, but ELOVL5 is intact in the patient
Parallel to the efforts to delineate the breakpoints, we quantified the amount of the transcript present in the patient for the genes mentioned above. As described in the “Methods” section, multiple reference genes have been used to normalize the data and RNA from 10 unrelated controls were used to assess the normal distribution of expression for CDK19 and ELOVL5 (Fig. 4a). The transcript level for CDK19 in the patient was found to be 54% (p = 0.022) when compared with the control mean, but that of ELOVL5 was not significantly reduced (Fig. 4b). This indicates to the most likely situation that one copy of the CDK19 has become non-functional (haploinsufficiency) due to the pericentric inversion at 6q21. To further refine the breakpoint on 6q21, overlapping fragments of ~10 kb were generated by long-range PCR from intron 1 of CDK19 and these products were used as FISH probes. Owing to the repeat-rich nature of the template, most of these probes did not give reproducible results. However, one probe (C″ in Fig. 2) shown complete translocation to the p-arm upon inversion (Fig. 2c) placing the break-point distal to this probe. This data along with the data from two BAC clones as mentioned above almost certainly puts the 6q21 inversion breakpoint within intron 1 of CDK19 confirming our initial observations (Fig. 2d). For 6p12.1, based on the transcript data, the breakpoint should be outside and proximal to ELOVL5. To confirm that no partial duplication of the ELOVL5 gene is involved with this inversion, we have performed multiplex ligation-dependent probe amplification (MLPA) and genomic QPCR in the region of the probes overlapping with ELOVL5 and none of the probes showed any copy number changes (data not shown).
Fig. 4.
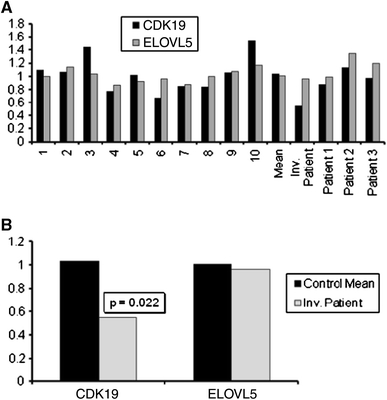
Relative quantitation of CDK19 and ELOVL5 transcripts. a For relative quantitation of the transcript amounts of CDK19 and ELOVL5 in the patient, the expression levels were compared with that of the mean of 10 controls (1–10). The graph also shows the mean value for the 10 controls and the four patients included in the study. Among these, the sample marked with ‘Inv. patient 1’ is the patient with the de novo pericentric inversion. b The quantity of CDK19 and ELOVL5 transcripts of the inversion patient was compared with the control mean (a). While the transcript amount for ELOVL5 was not significantly different and that for CDK19 was found to be only 54% compared to the controls, which was statistically significant (p = 0.022). The results indicated that the patient is haploinsufficient for the CDK19 gene. All quantitation was done using the 2ΔΔCt method after normalization with the geometric mean of the expression of GusB and RNF111 for reference genes as described in the “Methods” section. p value was calculated from the Z score derived from the ΔC t values for expression
Screening the CDK19 gene
The pericentric inversion event was de novo which most likely occurred in the germ line of one of the parents. To test whether the other CDK19 copy was involved, we sequenced the exons of this gene. However, no potentially pathologic sequence variant was identified in the open reading frame of CDK19. We ascertained nine additional patients with similar phenotypic features. All of them showed a normal karyotype, and no mutations were found in the CDK19 open reading frame, and the transcript levels were normal in three of the nine patients in which RNA levels could be tested (AM, FPMC, unpublished data). In 100 sporadic patients with mental retardation in which the Fragile-X gene has been excluded, we also screened the CDK19 gene for deletions using three MLPA probes covering the gene. None of the patients shows an abnormal copy number suggesting that haploinsufficiency of CDK19 is not a common cause of mental retardation.
The Drosophila orthologue of CDK19 is a regulator of dendritic development
Because the patient exhibits developmental anomalies, we examined the physiological role of a CDK19 orthologue in the complex developing nervous system of Drosophila melanogaster. Human CDK19 (NP_055891.1) is a paralogue of the CDK8 protein (NP_001251.1), and is also referred to as CDK8-like. The protein products of these two genes share 77% identity suggesting that they are functionally related. Drosophila contains a single (two-to-one) orthologue to both proteins, Cdk8 (NP_536735.2) (Leclerc et al. 1996), which shares 66% identity with human CDK19 and 71% identity with human CDK8. Construction of an unrooted phylogenetic tree using the neighbor-joining algorithm (http://www.ebi.ac.uk/clustalw) (Saitou and Nei 1987) shows that the Drosophila Cdk8 protein is almost equally related to both CDK19 and CDK8 (Fig. 5a). Furthermore, biochemical and genetic studies have confirmed a structural and functional conservation of Drosophila Cdk8 when compared with yeast and mammalian models (Loncle et al. 2007).
To examine the function of Cdk8 in fly development, we first performed a ubiquitous knockdown of this gene by driving expression of an UAS-Cdk8 RNAi transgene using the Act-Gal4 driver. Ubiquitous knockdown of cdk8 resulted in lethality at pupal and pharate adult stages. Analysis of the knock-down efficiency in third instar larvae showed that Cdk8 mRNA was significantly reduced by 68% (p < 0.05) when compared with controls (Fig. 5b) demonstrating that partial activity of this gene is not sufficient to achieve normal development. This is consistent with the phenotype observed in Cdk8 null mutant flies, which also show developmental lethality (Loncle et al. 2007) and the haploinsufficiency of this gene seen in our patient.
Because the neuronal pathology in our patient is uncertain, but dendritic abnormalities are commonly observed in MR (Kaufmann and Moser 2000), we examined whether Cdk8 affects dendritic development in the Drosophila type 4 md sensory neurons located in the peripheral nervous system of the Drosophila larvae. These neurons are amenable to quantitative analysis and have previously been shown to be relevant in other fly models for mental retardation (Lee et al. 2003; Lu et al. 2009). We found that knockdown of Cdk8 in Drosophila type 4 md neurons by expression of UAS-Cdk8 RNAi with the 477-Gal4 driver resulted in reduced dendritic branching and altered morphology of the dendritic arbour (Fig. 5c, d). Quantification of dendritic branching revealed a 35% reduction (p < 0.001) in dendrite ends upon Cdk8 knockdown (Fig. 5c), which appeared to be due in part to a loss of small dendritic protrusions of a length <10 μm (Fig. 5d). This trend was seen consistently and is highlighted in the lower panels of Fig. 5d where such protrusions (marked by red dots) are 35% less in mutant than in control neurons. A similar defect was also observed with a second UAS-Cdk8 RNAi line indicating that this phenotype does not result from non-specific effects due to insertion of the transgene. In addition, 90% of dendritic arbours examined for Cdk8 knockdown showed restricted extension and did not occupy the full field of view examined by confocal microscopy (Fig. 5d). In many cases, the unoccupied area was filled by a more faintly stained type 4 md neuron from the ventral cluster (shown in Fig. 5d) and in some cases no other dendrites were stained in the ‘unoccupied’ area (data not shown). In contrast, wild-type neurons always occupy nearly the entire field of view demonstrating a clear morphological defect in resulting from knockdown of the CDK19 orthologue in these neurons.
Discussion
In this article, we described that the microcephaly, congenital retinal fold and mild mental retardation is likely caused by CDK19 haploinsufficiency in a female Caucasian patient born to non-consanguineous parents. CDK19 expression was found in a wide variety of human tissues, with high expression in fetal brain and fetal retina (Fig. 3), the two tissues involved in the phenotypes observed in the patient. Furthermore, our functional analysis in Drosophila shows that the closest CDK19 orthologue, Cdk8, is a regulator of dendritic development, an important factor in the etiology of mental retardation (Kaufmann and Moser 2000). In addition, recent reports showed that loss of CDK8 in Drosophila disrupts the neural wiring of the eye and development of sensory bristles in the peripheral nervous system (Loncle et al. 2007; Berger et al. 2008), implying a role for Cdk8 both in dendritic development and in development of peripheral sensory organs including the eye.
Because Drosophila contains only one orthologue to the mammalian paralogues CDK19 and CDK8, analysis in this system is not confounded by potential redundancies between CDK19 and CDK8 that may occur in mammalian model systems and in humans. This is likely to contribute to the more dramatic phenotypes (e.g. lethality) in the model as compared to our patient.
CDK19 encodes a protein that is one of the components of the mediator co-activator complex. The mediator complex is a multi-protein complex required for transcriptional activation by DNA-binding transcription factors for genes transcribed by RNA polymerase II (pol II). The protein is 77% identical to cyclin-dependent kinase 8 (CDK8), which is another component of the mediator complex (Elmlund et al. 2006).
CDK8 (cyclin-dependent kinase 8), along with CycC, Med12, and Med13, form a repressive module (the Cdk8-submodule), which inhibits the transcriptional coactivator function of the mediator complex by prevention of interactions with pol II and can operate independently of the mediator (Elmlund et al. 2006; Knuesel et al. 2009a, b). Based on the homology with CDK8, it is reasonable to predict that CDK19 also has related functions. Interestingly, the CDK8-submodule of the mediator complex is linked to mental retardation through MED12, a causative gene for FG syndrome, a neurological disorder characterized by mental retardation, macrocephaly, hypotonia and constipation (Risheg et al. 2007). MED12 is known to regulate neuronal gene expression through cooperation with the neuronal gene silencing factor REST and euchromatin histone methyltransferase 2 (EHMT2/G9a) (Ding et al. 2008). EHMT2/G9a activity is dependent on interactions with its close homolog EHMT1 (Tachibana et al. 2005), which is involved in Kleefstra syndrome, a neurological disorder characterized by mental retardation and other morphological and behavioral phenotypes (Kleefstra et al. 2006). EHMT1 and EHMT2 have also been linked to REST transcriptional activity through interactions with the histone demethylase JARID1c (Tahiliani et al. 2007), yet another mental retardation gene with a known role in dendritic development (Iwase et al. 2007; Jensen et al. 2005). Taken together, these data strongly suggest that the protein complexes containing CDK19 form a network of transcriptional repressors that effect neuronal development and function and contribute to the etiology of mental retardation. However, a possibility cannot be excluded that in human and other mammals, CDK8 and CDK19 work in synergy, and lack of one molecule can be compensated by the other.
Another important cyclin-dependent kinase, CDKL5, has been associated with atypical forms of Rett syndrome (RS), an X-linked neurological disorder, characterized by microcephaly, seizures and mental retardation (Tao et al. 2004; Weaving et al. 2005). CDK19 has 32% identity and 49% similarity with CDKL5 at the amino acid level. The different phenotype described in this study and the eye phenotypes might be explained by the differences in spatial and temporal expression of the two genes and having different targets for phosphorylation. A large deletion in patients with X-linked retinoschisis (XLR, MIM 312700) was found to have disrupted at least the last coding exon of CDKL5, in addition to disruption of the RS1 gene (Huopaniemi et al. 2000). One of the patients had congenital nystagmus, which we also observed in our patient. Interestingly, XLRS patients also show splits and breaks in retina and occasionally retinal detachment, which are features of other vitreoretinopathies as well as overlaps with the features in the patient described in this study.
Recently, a growing number of studies have shown the involvement of ciliary proteins in diseases involving mental retardation (Badano et al. 2006). Some examples include Bardet–Biedl syndrome (BBS, MIM 209900), Meckel–Gruber syndrome (MKS1, MIM 249000 and MKS2, MIM 603194), and Joubert syndrome (MIM 610188). Bardet–Biedl and Joubert syndrome also manifest microcephaly and mental retardation in addition to retinal degeneration in some cases. We hypothesized that CDK19 might be present in the cilia and found it in the ciliary proteome database (Gherman et al. 2006). It was reported that the BBS patients have olfaction defects (anosmia) which has been suggested to be a general manifestation of ciliary protein defects (Kulaga et al. 2004). To test our hypothesis, we have performed the scratch-n-sniff test for our patient which revealed that she is highly anosmic (score 16 at the age of 41 years). However, this data can be confounded due to underdeveloped mental abilities of the patient.
In this article, we have described a female patient with microcephaly, congenital retinal fold and mild mental retardation, who has a pericentric inversion in one copy of chromosome 6, causing haploinsufficiency of CDK19 gene. The expression profile of CDK19, its homology with other serine/threonine kinases, and its role in the in Drosophila nervous system development, together strongly suggests CDK19 as an important determining factor for this syndrome.
Acknowledgments
The authors thank Drs. Arjan P.M. de Brouwer, Anneke I. den Hollander and Karin W. Littink for their help during various phases of the work. Dr. K. Devriendt from Leuven (Belgium) is acknowledged for providing us with DNA samples from six patients with similar phenotypes. Thanks also to Astrid Oudakker, Saskia D. van der Velde-Visser, Christel Beumer and Tannavi Kiran for expert technical assistance. This study was supported by European Union Research Training Network Grant RETNET MRTN-CT-2003-504003, a Vidi grant from The Netherlands Organization for Scientific Research (NWO) to AS and Council of Scientific and Industrial Research (CSIR), India funded project, “An Integrative Biology Approach in Deciphering Genotype–Phenotype Correlation for Human Complex Disorders” (SIP-0006).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
A. Mukhopadhyay and J. M. Kramer contributed equally to this work.
Contributor Information
Arijit Mukhopadhyay, Phone: +91-11-27666156, FAX: +91-11-27667471, Email: arijit@igib.res.in, Email: arijit@igib.in.
Frans P. M. Cremers, Phone: +31-24-3614017, FAX: +31-24-3668752, Email: f.cremers@antrg.umcn.nl
References
- Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- Berger J, Senti KA, Senti G, Newsome TP, Asling B, Dickson BJ, Suzuki T. Systematic identification of genes that regulate neuronal wiring in the Drosophila visual system. PLoS Genet. 2008;4:e1000085. doi: 10.1371/journal.pgen.1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- de Brouwer AP, van Bokhoven H, Kremer H. Comparison of twelve reference genes for normalisation of gene expression levels in EBV cell lines and fibroblasts. Mol Diag Ther. 2006;10:197–204. doi: 10.1007/BF03256458. [DOI] [PubMed] [Google Scholar]
- de Bruijn DR, Kater-Baats E, Eleveld M, Merkx G, van Kessel AG. Mapping and characterization of the mouse and human SS18 genes, two human SS18-like genes and a mouse Ss18 pseudogene. Cytogenet Cell Genet. 2001;92:310–319. doi: 10.1159/000056920. [DOI] [PubMed] [Google Scholar]
- Ding N, Zhou H, Esteve PO, Chin HG, Kim S, Xu X, Joseph SM, Friez MJ, Schwartz CE, Pradhan S, Boyer TG. Mediator links epigenetic silencing of neuronal gene expression with X-linked mental retardation. Mol Cell. 2008;31:347–359. doi: 10.1016/j.molcel.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmlund H, Baraznenok V, Lindahl M, Samuelsen CO, Koeck PJ, Holmberg S, Hebert H, Gustafsson CM. The cyclin-dependent kinase 8 module sterically blocks mediator interactions with RNA polymerase II. Proc Natl Acad Sci USA. 2006;103:15788–15793. doi: 10.1073/pnas.0607483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner S. Congenital retinal folds and microcephaly. Arch Ophthalmol. 1941;25:93–100. [Google Scholar]
- Gherman A, Davis EE, Katsanis N. The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nat Genet. 2006;38:961–962. doi: 10.1038/ng0906-961. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development. 2002;129:2867–2878. doi: 10.1242/dev.129.12.2867. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell. 2003;112:805–818. doi: 10.1016/S0092-8674(03)00160-0. [DOI] [PubMed] [Google Scholar]
- Huopaniemi L, Tyynismaa H, Rantala A, Rosenberg T, Alitalo T. Characterization of two unusual RS1 gene deletions segregating in Danish retinoschisis families. Hum Mutat. 2000;16:307–314. doi: 10.1002/1098-1004(200010)16:4<307::AID-HUMU3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Jarmas AL, Weaver DD, Ellis FD, Davis A. Microcephaly, microphthalmia, falciform retinal folds, and blindness: a new syndrome. Am J Dis Child. 1981;135:930–933. doi: 10.1001/archpedi.1981.02130340036013. [DOI] [PubMed] [Google Scholar]
- Jensen LR, Amende M, Gurok U, Moser B, Gimmel V, Tzschach A, Janecke AR, Tariverdian G, Chelly J, Fryns JP, et al. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am J Hum Genet. 2005;76:227–236. doi: 10.1086/427563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann WE, Moser HW. Dendritic anomalies in disorders associated with mental retardation. Cereb Cortex. 2000;10:981–991. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- Kleefstra T, Brunner HG, Amiel J, Oudakker AR, Nillesen WM, Magee A, Geneviève D, Cormier-Daire V, van Esch H, Fryns JP, Hamel BC, Sistermans EA, de Vries BB, van Bokhoven H. Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am J Hum Genet. 2006;79:370–377. doi: 10.1086/505693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ. The human CDK8 subcomplex is a molecular switch that controls mediator coactivator function. Genes Dev. 2009;23:439–451. doi: 10.1101/gad.1767009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuesel MT, Meyer KD, Donner AJ, Espinosa JM, Taatjes DJ. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Mol Cell Biol. 2009;29:650–661. doi: 10.1128/MCB.00993-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulaga HM, Leitch CC, Eichers ER, Badano JL, Lesemann A, Hoskins BE, Lupski JR, Beales PL, Reed RR, Katsanis N. Loss of BBS proteins causes anosmia in humans and defects in olfactory cilia structure and function in the mouse. Nat Genet. 2004;36:994–998. doi: 10.1038/ng1418. [DOI] [PubMed] [Google Scholar]
- Leclerc V, Tassan JP, O’Farrell PH, Nigg EA, Leopold P. Drosophila Cdk8, a kinase partner of cyclin C that interacts with the large subunit of RNA polymerase II. Mol Biol Cell. 1996;7:505–513. doi: 10.1091/mbc.7.4.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Li W, Xu K, Bogert BA, Su K, Gao FB. Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development. 2003;130:5543–5552. doi: 10.1242/dev.00792. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Loncle N, Boube M, Joulia L, Boschiero C, Werner M, Cribbs DL, Bourbon HM. Distinct roles for Mediator Cdk8 module subunits in Drosophila development. EMBO J. 2007;26:1045–1054. doi: 10.1038/sj.emboj.7601566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Wang F, Li Y, Ferris J, Lee JA, Gao FB. The Drosophila homologue of the Angelman syndrome ubiquitin ligase regulates the formation of terminal dendritic branches. Hum Mol Genet. 2009;18:454–462. doi: 10.1093/hmg/ddn373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda Y. Two cases of ablatio falciformis congenitaand two other cases of ocular congenital anomalies, which appeared in a pedigree with consanguineous marriages. Jpn J Clin Ophthalmol. 1962;16:325–331. [PubMed] [Google Scholar]
- Nguyen TN, Der Kaloustian VM, Barsoum-Homsy M, Dembinska O, Koenekoop RK. Congenital microcephaly, juvenile retinal dystrophy and normal mentation in a mildly dysmorphic child. Can J Ophthalmol. 2005;40:195–199. doi: 10.1016/S0008-4182(05)80034-1. [DOI] [PubMed] [Google Scholar]
- Nishimura M, Yamana T, Sugino M, Kohno T, Yamana Y, Minei M, Sanui H. Falciform retinal fold as sign of familial exudative vitreoretinopathy. Jpn J Ophthalmol. 1983;27:40–53. [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger E. Microcephalie und microphthalmie. Archiv fur Augenheilk. 1885;14:1–11. [Google Scholar]
- Risheg H, Graham JM, Jr, Clark RD, Rogers RC, Opitz JM, Moeschler JB, Peiffer AP, May M, Joseph SM, Jones JR, et al. A recurrent mutation in MED12 leading to R961W causes Opitz–Kaveggia syndrome. Nat Genet. 2007;39:451–453. doi: 10.1038/ng1992. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3–K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, Li J, Rao A, Shi Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- Tao J, van Esch H, Hagedorn-Greiwe M, Hoffmann K, Moser B, Raynaud M, Sperner J, Fryns JP, Schwinger E, Gecz J, et al. Mutations in the X-linked cyclin-dependent kinase-like 5 (CDKL5/STK9) gene are associated with severe neurodevelopmental retardation. Am J Hum Genet. 2004;75:1149–1154. doi: 10.1086/426460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nouhuys CE. Congenital retinal fold as a sign of dominant exudative vitreoretinopathy. Graefes Arch Ophthalmol. 1981;217:55–67. doi: 10.1007/BF00410881. [DOI] [PubMed] [Google Scholar]
- van Nouhuys CE. Dominant exudative vitreoretinopathy and other vascular developmental disorders of the peripheral retina. Doc Ophthalmol. 1982;54:1–414. doi: 10.1007/BF00681132. [DOI] [PubMed] [Google Scholar]
- van Nouhuys CE. Autosomal recessive vitreoretinopathy and encephaloceles. Am J Ophthalmol. 1982;94:820. doi: 10.1016/0002-9394(82)90316-6. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, vanRoy N, dePaepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034 [DOI] [PMC free article] [PubMed]
- Wall FE, Henkel RD, Stern MP, Jenson HB, Moyer MP. An efficient method for routine Epstein-Barr virus immortalization of human B lymphocytes. In Vitro Cell Dev Biol Anim. 1995;31:156–159. doi: 10.1007/BF02633976. [DOI] [PubMed] [Google Scholar]
- Weaving LS, Ellaway CJ, Gecz J, Christodoulou J. Rett syndrome: clinical review and genetic update. J Med Genet. 2005;42:1–7. doi: 10.1136/jmg.2004.027730. [DOI] [PMC free article] [PubMed] [Google Scholar]


