Figure 3. RNase P RNA Binds to PNPASE and May Function in PNPASE-Dependent tRNA Processing.
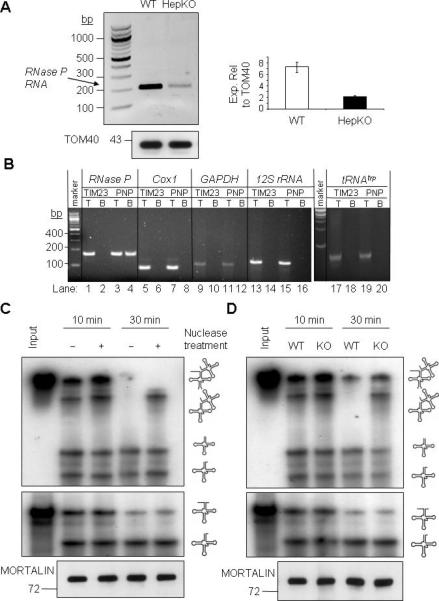
(A) Left- RNA was isolated from WT and HepKO liver mitochondria following nuclease treatment. RT-PCR was performed with primers that amplify nuclear-encoded RNase P RNA (212-bp). Right- QPCR analysis of RNase P RNA expression relative to TOM40 protein in isolated mitochondria.
(B) PNPASE-HisPC (PNP) or TIM23-HisPC (TIM23) was purified from stably-transfected HEK293 cells. Candidate interacting RNAs that co-purified in the final eluate with PNPASE-HisPC and TIM23-HisPC were identified by primer-specific RT-PCR. T is the total lysate (0.3% of the reaction) before mitochondrial purification and B is the bound fraction. Note that only RNase P RNA bound to PNPASE-HisPC (lane 4).
(C) Mitoplast extract (10 μg) was prepared from nuclease-treated, intact WT liver mitochondria. The extract was then treated with nuclease (+), as indicated, and then inactivated with EDTA and EGTA. The nuclease-treated or untreated extract was incubated with abutted tRNAs (tRNAHistRNASer) or a single tRNA (tRNALys) at 25°C for 10 or 30 min. RNA was separated on an urea-acrylamide gel and detected by autoradiography. A MORTALIN immunoblot shows equivalent mitoplast extract in each assay.
(D) Mitoplast extract was prepared from nuclease-treated, intact WT or HepKO liver mitochondria. The enzymatic assay was performed as described for panel C. See also Figure S5.
