Figure 5. PNPASE Mutations that Inactivate RNA Processing do not Affect RNA Import or Stability.
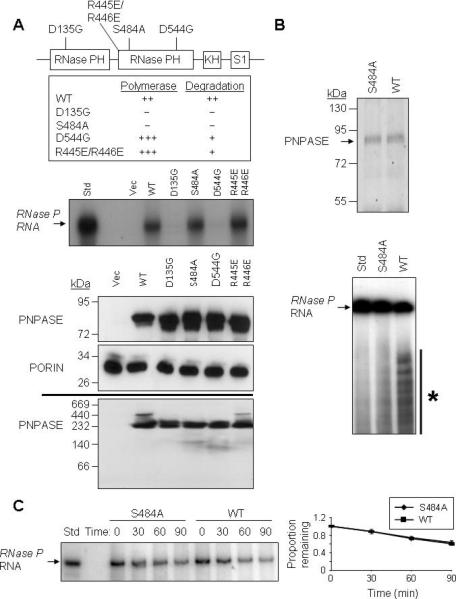
(A) Upper- Schematic for the positions of point mutations made in the PNPASE protein. Listed are the in vitro effects of mutations on 3’ polymerase and RNA degrading activities, from (Portnoy et al., 2008). Middle- Import reactions were performed as in Figure 4A. Radiolabeled RNase P RNA was incubated with isolated yeast mitochondria expressing an empty vector or the listed PNPASE constructs. Lower panels- Immunoblot of WT and point mutant PNPASE yeast transfectants used in panel A import assay. A PORIN immunoblot confirms the co-localization of PNPASE WT and mutants in yeast mitochondria. The assembly state of WT and point mutant PNPASE was determined by solubilization with 1% digitonin and separation on a 6-16% BN gel, followed by PNPASE immunoblot.
(B) Upper- WT and S484A PNPASE IPs from yeast mitochondria were used to analyze RNA degradation properties. Lower- WT or S484A mutant PNPASE was incubated with radiolabeled RNase P RNA for 10 min at 25°C to assess degradation activity. The asterisk marks degradation products.
(C) Left- Following in vitro import of radiolabeled RNase P RNA and nuclease treatment to remove non-imported RNA, mitochondria were incubated for up to 90 min at 25°C and aliquots removed at the indicated time points. The RNA was then resolved by urea-acrylamide gel electrophoresis. Right- RNase P RNA that was not degraded was quantified using FX imager; n = 3.
