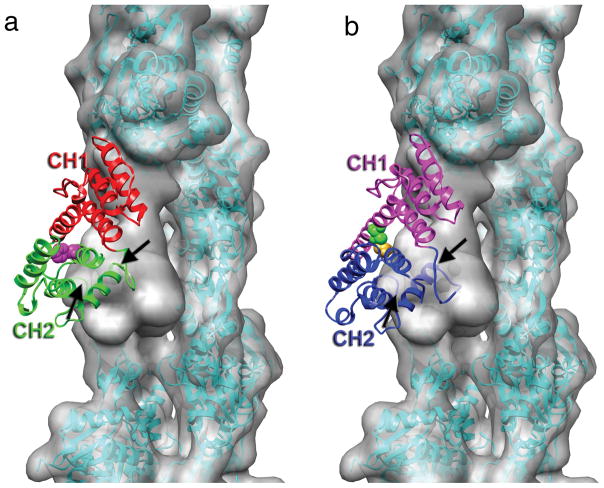
Figure 3.
Steric clash between calponin homology domain 2 (CH2) of α-actinin and actin. (a) The crystal structure of the actin-binding domain (ABD) of α-actinin (PDB 1TJT) has been docked onto F-actin so that CH1 is superimposed on the model for CH1 bound to F-actin in Fig. 2a. CH2 has a significant penetration into the actin volume, indicated by the black arrows. The position of residue 255 at the CH1-CH2 interface is indicated by the magenta space-filling atoms. The K255E mutation destabilizes this interface, and increases the affinity of α-actinin for actin14. (b) The binding of α-actinin’s ABD to F-actin may likely explain observations made about plectin19, another actin-binding protein containing tandem CH domains. When CH1 of plectin (PDB 1MB8) is aligned with CH1 of α-actinin, CH2 of plectin makes a steric clash with an actin protomer (black arrow). When the two plectin CH domains can separate, the affinity for F-actin is approximately three times greater than when the two CH domains are crosslinked (the disulfide crosslink is indicated by the space filling atoms at the CH1-CH2 interface) in a closed conformation19, suggesting that strain introduced into either F-actin or plectin to eliminate the steric clash reduces the binding affinity.