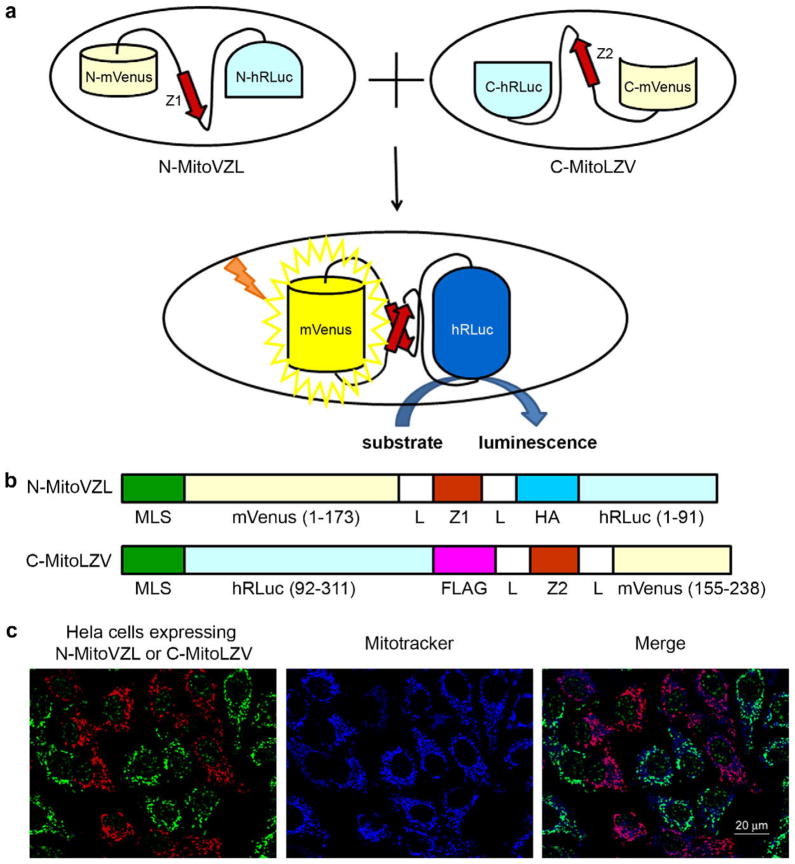
Fig. 1.
Mitochondrial fusion quantification via split-Renilla luciferase complementation. (A) Principle of split-Renilla luciferase complementation to quantitate mitochondrial fusion. Upon fusion of two mitochondria individually expressing a hybrid protein encoding half of the split-Renilla luciferase, the antiparallel leucine zipper pair (red) pulls the constructs together and the luciferase fragments reconstitute and generate luminescence in the presence of substrate. The split-Venus also reconstitutes and generates yellow fluorescence. (B) Schematic representations of domain structures of split-Renilla luciferase. Both constructs have a mitochondrial localization sequence (MLS) in their N-termini. The N-terminal monomeric (m) Venus fragment is fused to the N-terminal hRLuc fragment with a leucine zipper peptide (Z1) in the middle flanked by two linkers (L) and an HA peptide. The C-terminal hRLuc fragment is fused with the C-terminal Venus fragment and the corresponding antiparallel leucine zipper peptide (Z2) flanked by the two same linkers (L) and a FLAG peptide. (C) Mitochondrial localization of split-Renilla luciferase constructs. HeLa cells expressing each construct were mixed and stained for the HA (green) and FLAG (red) epitopes built into the fusion proteins. Note that the anti-HA antibody non-specifically detects a speckled nuclear protein present in all cells. The mitochondria were stained with MitoTracker far red 633 (blue). Scale bar, 20 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)