Figure 5.
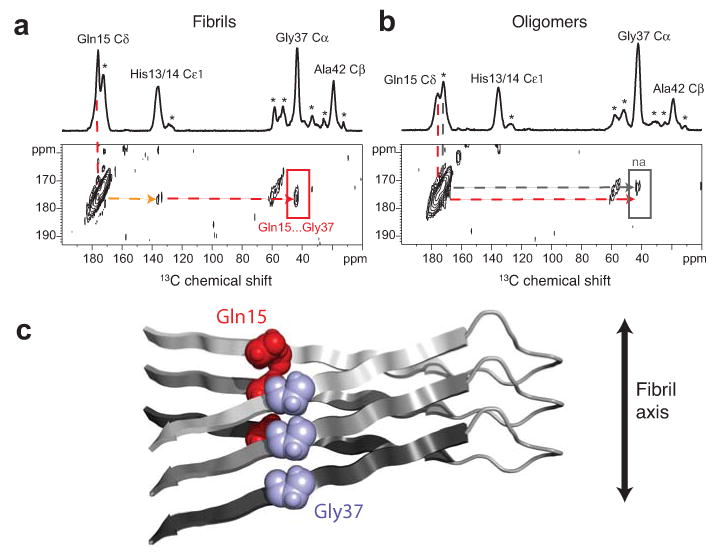
β-strands are staggered in Aβ42 fibrils. (a) Above, one dimensional 13C-spectrum showing chemical shift assignments for His13/14, Gln15, Gly37 and Ala42 in Aβ42 fibrils formed from an equimolar mixture of Aβ42-HQA:Aβ42-G37 peptides. Natural abundance 13C assignments are marked with an asterisk. Below, region of the two dimensional DARR NMR spectrum showing specific 13C…13C inter-molecular contacts between Gln15 and Gly37 (red arrow), intra-molecular contacts between Gln15 and His13/14 (orange arrow), and no contact between Gln15 and Ala42, indicating a staggered, domain-swapped architecture. (b) Model of staggering between the N- and C-terminal β-strands at the Gln15-Gly37 contact in Aβ42 fibrils. (c) Above, one dimensional 13C-spectrum showing chemical shift assignments for His13/14, Gln15, Gly37 and Ala42 in Aβ42 oligomers formed from an equimolar mixture of Aβ42-HQA:Aβ42-G37 peptides. Below, two dimensional DARR NMR spectrum showing no molecular contacts between Gln15 and His13/14, Gly37, or Ala42 (red arrow), indicating the absence of a staggered, domain swapped architecture in Aβ42 oligomers. Only a small natural abundance (na) cross-peak is observed (gray arrow).
