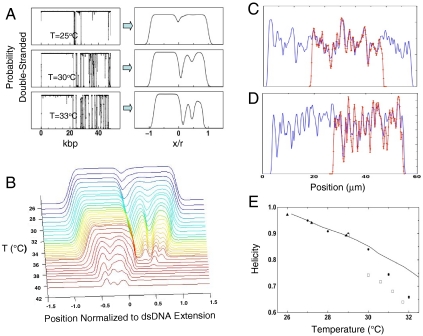
Fig. 4.
Theoretical prediction of barcode structure from dsDNA melting probability (A). Theoretical melting traces can be obtained directly from melting-probability calculations by assuming values for ds/ss DNA stretching, fluorescence, and convolving the profile with a Gaussian to simulate optical broadening. (B) Evolution of λ-phage melting traces with temperature across the melting transition: by scanning temperature, both high AT and high GC regions are imaged. Details in high AT regions are visible at low temperature, wheras at higher temperature details in high GC regions can be imaged. (C) Experimental barcode profile (red) for single BAC RP11-125C7 molecule aligned to theoretical estimate of RP11-125C7 barcode (blue). (D) Experimental barcode profile (red) for single T4GT7 molecule (red) aligned to theoretical estimate of T4GT7 barcode (blue). The theoretical estimates for T4GT7 and BAC RP11-125C7 are created from two identical and adjoining sequences to capture trace permutation. (E) Fitted λ-phage helicities vs. temperature; circles, open squares, and triangles indicate measurements with different chips/setups. The variation between experimental runs arises primarily from variations in thermocouple calibration (∼1 °C error). The bold curve is a theoretical estimate of the helicity at 5 mM NaCl.