Fig. 1.
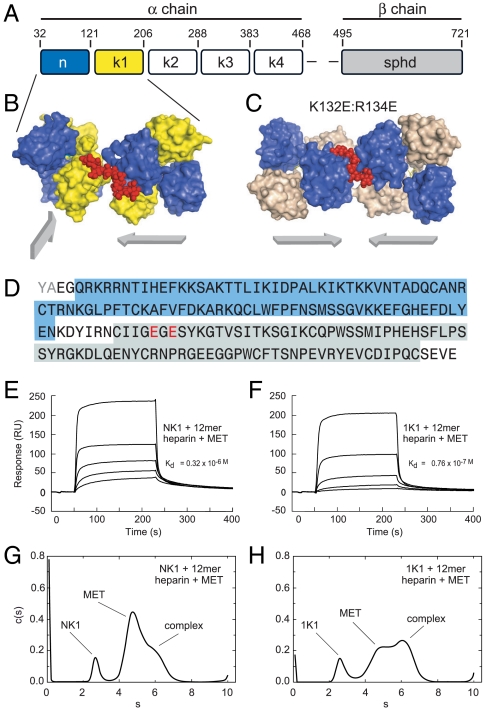
Structural features of the 1K1-heparin complex and interaction with the MET receptor (A) Domain structure of full length multidomain HGF/SF. The α-chain consists of N-terminal domain (amino acids 32–121) and four kringle domains(K1, K2, K3, and K4) and the β-chain contains serine protinease domain (spdh domain). (B) Crystal structure of NK1-heparin complex (16) (PDB accession 1GMO). Two NK1 dimers are shown bridged by heparin. N and K domains are shown in blue and yellow, heparin is shown in red (C) Crystal structure of 1K1-heparin complex (PDB accession 3MPK). Two 1K1 dimers are shown bridged by heparin. N and K domains are shown in blue and gray, heparin is shown in red. The figures in B and C have been drawn with Pymol. Reverse mutations K132E and R134E were introduced into K1 domain of 1K1 to inactivate the low-affinity heparin-binding sites. (D) and (E). (D) Amino acid sequence of 1K1 demonstrating the mutated sites. Binding of NK1 (E) and 1K1 (F) to MET567 in the presence of 12mer heparin using surface plasmon resonance. Twofold dilutions of each protein were used from a concentration of top concentration of 200 nM. The concentration of 12mer heparin in the sample and in the reaction buffer was 10 μM. (G) and (H). Velocity sedimentation analysis of ternary complexes of NK1-heparin-MET567 (G) and 1K1-heparin-MET567 (H). Data show plots of c(s) against s*20,w. In the presence of 10mer heparin, the amount of ternary complex is significantly higher for 1K1 than for NK1.